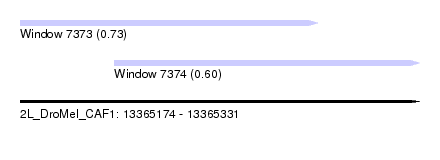
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,365,174 – 13,365,331 |
| Length | 157 |
| Max. P | 0.730636 |

| Location | 13,365,174 – 13,365,291 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
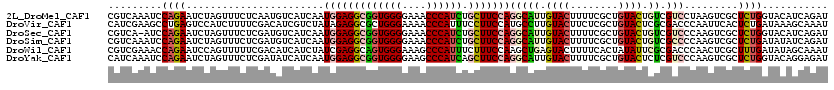
>2L_DroMel_CAF1 13365174 117 + 22407834 GGCUGCUCCUCCUCGGCGUUGACCUCGUCAUCC---UCCUCGUCAAAUCCAGAAUCUAGUUUCUCAAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUU .((((........)))).(((((..........---.....)))))....((((......))))(((((((.....(((((((((((((....)))).))))))))))))))))...... ( -37.26) >DroSec_CAF1 6055 116 + 1 GGCUGCUCCUCCUCGGCGUUGACCUCGUCAUCC---UCCUCGUCA-AUCCAGAAUCUAGUUUCUCGAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUU .((((........))))((((((..........---.....))))-))..((((......))))(((((((.....(((((((((((((....)))).))))))))))))))))...... ( -37.76) >DroSim_CAF1 5210 117 + 1 GGCUGCUCCUCCUCGGCGUUGACCUCGUCAUCC---UCCUCGUCAAAUCCAGAAUCUAGUUUCUCGAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUU .(((..........(((.(((((..........---.....)))))(((.((((......)))).)))))).....(((((((((((((....)))).)))))))))))).......... ( -37.16) >DroWil_CAF1 36733 120 + 1 GAUGACUUCUUUUCUUCGUCCUCUUCGUCAUCCAUUUCCUCGUCGAAACCAGAAUCCAGUUUUUCGACAUCAUCUAUCGAGGCAGUGGGAAAGCCCAUUUCUUUCCAAGCUGAGUACUUU ((((((....................)))))).........(((((((.............))))))).(((.((...((((.((((((....)))))).))))...)).)))....... ( -23.77) >DroYak_CAF1 5751 117 + 1 GGCUGCUCCUCCUCGGCGUUGCCCUCGUCAUCC---UCCUCAUCAAAUCCAGAAUCUAGUUUCUCGAUAUCAUCAAUGGAGGCGGUGGGGAAGCCCAUCAGCUUCCAGGCAUUGUACUUU (((.((.((.....)).)).)))...(((....---..........(((.((((......)))).)))........(((((((((((((....)))))).)))))))))).......... ( -37.30) >consensus GGCUGCUCCUCCUCGGCGUUGACCUCGUCAUCC___UCCUCGUCAAAUCCAGAAUCUAGUUUCUCGAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUU ...((((.......((((.......)))).................(((.((((......)))).)))........(((((((((((((....)))))).)))))))))))......... (-26.22 = -26.70 + 0.48)

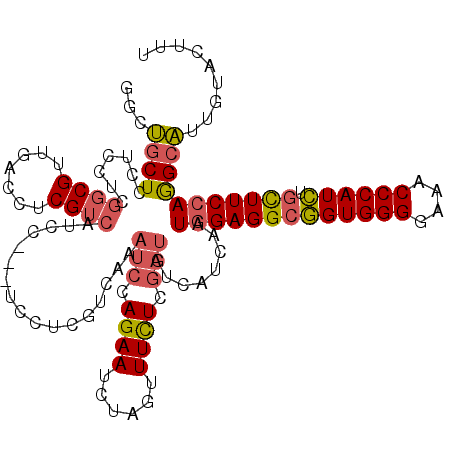
| Location | 13,365,211 – 13,365,331 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -23.33 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13365211 120 + 22407834 CGUCAAAUCCAGAAUCUAGUUUCUCAAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUUUCGCUGUACUGUCGUCCUAAGUCGCUCUGGUACAUCAGAU .........(((.....(((....(((((((.....(((((((((((((....)))).)))))))))))))))).))).....)))..(((..((((..((...))..)).))..))).. ( -41.00) >DroVir_CAF1 6967 120 + 1 CAUCGAAGCCUGAGUCCAUCUUUUCGACAUCGUCUAUAGAGGCGCUGGGAAAACCCAUUUCCUUCCAUGCCUUGUACUUCUCGCUGUACUCGCGACCCAAUUCACUCUGAUAAAGCAAAU ..((((((...((.....)).))))))..((((.....((((((..((((((.....))))))....))))))((((........))))..))))......................... ( -23.00) >DroSec_CAF1 6092 119 + 1 CGUCA-AUCCAGAAUCUAGUUUCUCGAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUUUCGCUGUACUGUCGUCCCAAGUCGCUCUGGUACAUCAGAU .....-...(((.....(((....(((((((.....(((((((((((((....)))).)))))))))))))))).))).....)))..(((..((.(((........))).))..))).. ( -42.30) >DroSim_CAF1 5247 120 + 1 CGUCAAAUCCAGAAUCUAGUUUCUCGAUGUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUUUCGCUGUACUGUCGCCCCAAGUCGCUCUGAUAUAUCAGAU .(((......((((......)))).(((((.((((.(((((((((((((....)))).)))))))))(((((.((((........)))).)).)))...........))))))))).))) ( -42.00) >DroWil_CAF1 36773 120 + 1 CGUCGAAACCAGAAUCCAGUUUUUCGACAUCAUCUAUCGAGGCAGUGGGAAAGCCCAUUUCUUUCCAAGCUGAGUACUUUUCACUAUAUUCGCGACCCAACUCGCUUUGAUAUAGCAAAU .(((((((.............)))))))......((((((((((((.((((((.......))))))..((.(((((..........)))))))......))).)))))))))........ ( -28.02) >DroYak_CAF1 5788 120 + 1 CAUCAAAUCCAGAAUCUAGUUUCUCGAUAUCAUCAAUGGAGGCGGUGGGGAAGCCCAUCAGCUUCCAGGCAUUGUACUUUUCGCUGUACUCUCGUCCCAAGUCGCUCUGGUACAGGAGAU ......(((.((((......)))).)))........(((((((((((((....)))))).)))))))...........((((.(((((((..((.(....).))....))))))))))). ( -36.90) >consensus CGUCAAAUCCAGAAUCUAGUUUCUCGAUAUCAUCAAUGGAGGCGGUGGGGAAACCCAUCUGCUUCCAGGCAUUGUACUUUUCGCUGUACUCUCGUCCCAAGUCGCUCUGAUACAGCAGAU .........((((.......................(((((((((((((....)))))).)))))))(((((.((((........)))).)).))).........))))........... (-23.33 = -24.00 + 0.67)
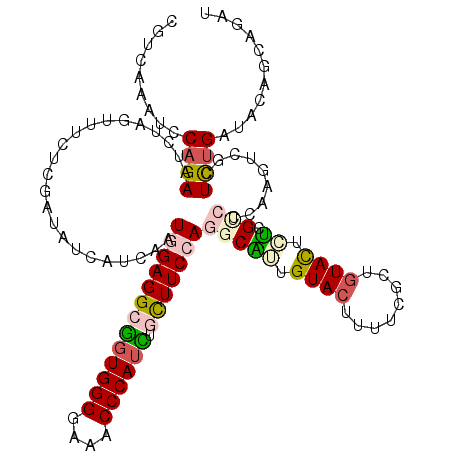
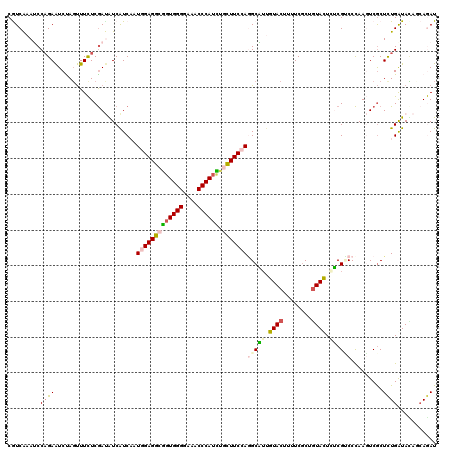
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:24:02 2006