| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,361,324 – 13,361,440 |
| Length | 116 |
| Max. P | 0.527499 |

| Location | 13,361,324 – 13,361,440 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -41.22 |
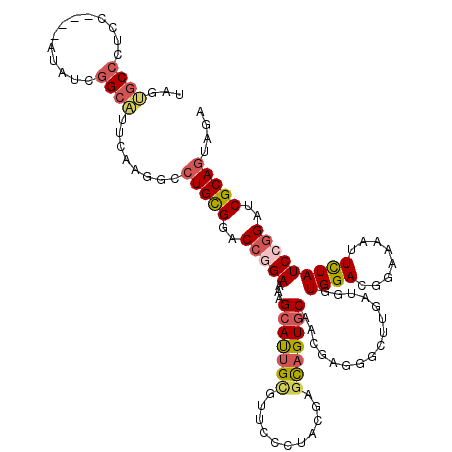
| Consensus MFE | -24.31 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
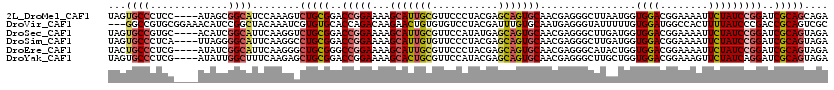
>2L_DroMel_CAF1 13361324 116 - 22407834 UAGUGCCCUCC----AUAGCGGCAUCCAAAGUCUGCGGACCGGAAAAGCAUUGCGUUCCCUACGAGCAGUGCAACGAGGGCUUAAUGGUGGACGGAAAAUUCUAUCCGGAUCGCAGCAGA ..(((((((..----..)).))))).....(.((((((.(((((...(((((((...........)))))))..((...(((....)))...))..........))))).)))))))... ( -37.20) >DroVir_CAF1 975 117 - 1 ---GGCCGUGCGGAAACAUCCGGCUACAAAUCGUGUGCACCAGACAAGAACUGUGUGUCCUACGAUUUGUGCAAUGAGGGUAUUUUUGUGGAUGGCCACUUUUAUCCCGACCGCAGUCGC ---(((((((((((((.((((.((.((((((((((..((((((.......))).)))..).))))))))))).....)))).)))))))..))))))..........((((....)))). ( -43.30) >DroSec_CAF1 2137 116 - 1 UAGUGCCGUGC----ACAUCGGCAUUCAAGGUCUGCGGACCGGAAAAGCAUUGCGUUCCAUAUGAGCAGUGCAACGAGGGCUUGAUGGUGGACGGAAAAUUCUAUCCGGAUCGCAGUAGA .(((((((...----....)))))))......((((((.(((((...(((((((...........)))))))....((((.((..((.....))..)).)))).))))).)))))).... ( -39.20) >DroSim_CAF1 1257 116 - 1 UAGUGCCCUCA----UUAGGGGCAUUCAAGGCCUGCGGACCGGAAAAGCAUUGUGUUCCCUACGAGCAGUGCAACGAGGGCUUGAUGGUGGACGGAAAAUUCUAUCCGGAUCGCAGUAGA .((((((((..----...))))))))......((((((.(((((......((.(((((((..(((((.(.....)....)))))..)).))))).)).......))))).)))))).... ( -41.12) >DroEre_CAF1 10557 116 - 1 UACUGCCCUCG----AUAUCGGCAUUCAAGGGCUGCGGGCCGGAAAAGCAUUGCGUUCCCUACGAGCAGUGCAACGAGGGCAUACUGGUGGACGGAAAAUUCUAUCCGGAUCGCAGUAGA ...((((((((----...(((((...((.....))...)))))....(((((((...........)))))))..))))))))(((((.(((.((((........))))..)))))))).. ( -44.90) >DroYak_CAF1 1347 116 - 1 UAGUGCCCUCG----AUAUUGGCUUUCAAGAGCUGCGGACCGGAAAAGCACUGCGUUCCAUACGAGCAGUGCAACGAGGGCUUGCUGGUGGACGGAAAGUUCUAUCAGGAUCGCAGUAGA ..(((((((((----.....(((((....))))).............(((((((...........)))))))..)))))))...(((((((((.....).))))))))....))...... ( -41.60) >consensus UAGUGCCCUCC____AUAUCGGCAUUCAAGGCCUGCGGACCGGAAAAGCAUUGCGUUCCCUACGAGCAGUGCAACGAGGGCUUGAUGGUGGACGGAAAAUUCUAUCCGGAUCGCAGUAGA ...((((.............))))........(((((..(((((...(((((((...........)))))))................((((........)))))))))..))))).... (-24.31 = -25.00 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:57 2006