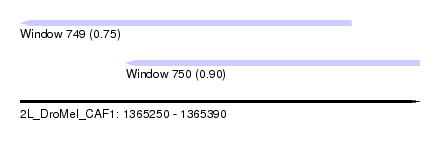
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,365,250 – 1,365,390 |
| Length | 140 |
| Max. P | 0.897662 |

| Location | 1,365,250 – 1,365,366 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -24.08 |
| Energy contribution | -25.05 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1365250 116 - 22407834 AUAACCAGGAUAACAACAAACGGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUAACCCCUCCCGGGCCCG---CGACCCUCC .....................((((.....((.((((((((((((.........)))))...))))))))).((.(((.(((................))).))).)---)).)))... ( -39.69) >DroPse_CAF1 172360 114 - 1 CCAACCCGCAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUUCCCGGUGGCCUGCCCCAUCCAAU----- .....((((....((((.....(.....).((.((((((((((((.........)))))...))))))))).))))..))))............((.((....)))).......----- ( -37.40) >DroEre_CAF1 140676 111 - 1 AUAACCAGGAUAACAACAAACGGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUAGCCCCGCCCCCGAACC---CGCC----- ....................((((..(((.((.((((((((((((.........)))))...))))))))).)))((((.((...........))))))))))....---....----- ( -37.60) >DroWil_CAF1 174008 99 - 1 AUAAC--------CAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUG---CACUUGGCUAGGAUAUAUCCUGCUU-GCCACCGCCUU---UGCU----- .....--------.........((((((..((..(((((((((((.........)))))...)))))---)...(((((((((....)))))...-))))..)).))---))))----- ( -32.80) >DroAna_CAF1 148384 111 - 1 AUAACCAGGAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUUGCCACUCCCUCCCCUC---UAUC----- .......(((....................((.((((((((((((.........)))))...)))))))))(((.((((.(((....))))))))))...)))....---....----- ( -31.50) >DroPer_CAF1 181267 114 - 1 CCAACCCGCAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUUCCCGGUGCCCUGCCCUAUCCAAU----- ......................(((((((.((.((((((((((((.........)))))...))))))))).)))((((((((.......)))..))))).)))).........----- ( -38.40) >consensus AUAACCAGGAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCGCGGAUAUAUCUUGCCCCUCCCCCGCCCC___CAAC_____ .....................((((...(((((((((((((((((.........)))))...)))))))..)))))....(((....)))............))))............. (-24.08 = -25.05 + 0.97)


| Location | 1,365,287 – 1,365,390 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -23.60 |
| Energy contribution | -24.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1365287 103 - 22407834 UUGGCUGGCUC----CUU-----GGCUG--CA-G-CCAUAACCAGGAUAACAACAAACGGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ...((..((((----(((-----(((((--((-(-((.......(.....)......(((((.....))))).))))))....))))))..(..((....))..))).)))..)). ( -42.30) >DroPse_CAF1 172395 110 - 1 UUUGGUGGCCCUCCCCGC-----CGCUCCUCA-GACACCAACCCGCAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ...(((((......))))-----)........-....(((((..((.............(((.....)))((((((((((((.........)))))...))))))))).))))).. ( -36.30) >DroWil_CAF1 174039 94 - 1 UUC----UUUU----CUUCGUUUUUUUA--CACG-CCAUAAC--------CAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUG---CACUUGGCU ...----....----.............--...(-(((....--------.........(((.....))).(((((((((((.........)))))...)))))---)...)))). ( -24.80) >DroYak_CAF1 125779 94 - 1 ---------UC----CUU-----GGCUG--CA-G-CCAUAACCAGGAUAACAACAAACGGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ---------((----(((-----(((..--..-)-))).....)))).............((.(((.((.((((((((((((.........)))))...))))))))).))).)). ( -34.60) >DroAna_CAF1 148416 100 - 1 UUG----GUGC----CUU-----GGCUC--CA-UCCCAUAACCAGGAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ..(----(.((----...-----.)).)--)(-(((........))))............((.(((.((.((((((((((((.........)))))...))))))))).))).)). ( -35.50) >DroPer_CAF1 181302 110 - 1 UUUGGUGGCCCUCCCCAC-----CGCUCCUCA-GACCCCAACCCGCAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ...(((((......))))-----)........-....(((((..((.............(((.....)))((((((((((((.........)))))...))))))))).))))).. ( -36.50) >consensus UUG____GCUC____CUU_____GGCUC__CA_G_CCAUAACCAGGAUAACAACAAACAGGCAAACAGCUCGCGGCUGCCACGAGCCAAAAGUGGCAUCAGCUGCGGCAGUUGGCG ............................................................((.(((.((.((((((((((((.........)))))...))))))))).))).)). (-23.60 = -24.27 + 0.67)

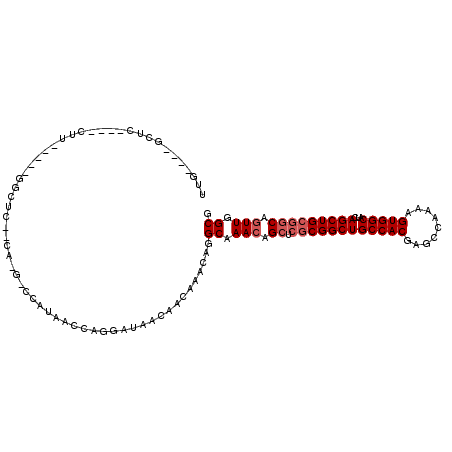
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:51 2006