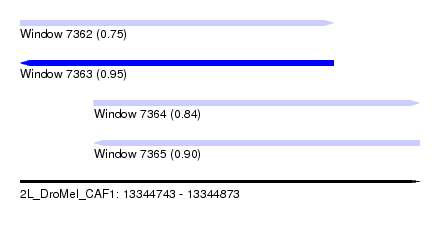
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,344,743 – 13,344,873 |
| Length | 130 |
| Max. P | 0.948564 |

| Location | 13,344,743 – 13,344,845 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13344743 102 + 22407834 UUGGAGUCGAGAGUGCGC---CCGGCC--ACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGUACAAAUGUACAAAUGUACUCCGGUG (..(((.((((.((((..---..((..--.))...((((....))))...)))).)))).)))..)((....(((((..(((((....)))))..)))))....)). ( -29.10) >DroSec_CAF1 57028 96 + 1 UUGGAGUCGAGAGUGCGC---CCGGCCUCACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU--------ACAAUUGUACUCCGGUG (..(((.((((.((((..---..((.....))...((((....))))...)))).)))).)))..)((((.(((((....))--------))).))))......... ( -27.10) >DroSim_CAF1 57369 96 + 1 UUGGAGCCGGGAGUGCGC---GCGGCCUCACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU--------ACAAAUGUACUCCGGUG .....((((((((((..(---(.((.....)).).((((....))))...)..)))))........(((..(((((....))--------)))..)))...))))). ( -26.90) >DroYak_CAF1 57602 105 + 1 UUGGAGUCGAGAGUGCGCCUCCUGGCC--ACCUCAUAUGCUCGCAUACGUGUACACUCGCCUCUGAACGAAUGUACAAAUGUAGAAAUGUACAAAUGUACUCCAGUG (((((((((((.(((.(((....))))--)))))((((..((((((...((((((.(((........))).)))))).)))).)).))))......).))))))).. ( -34.50) >consensus UUGGAGUCGAGAGUGCGC___CCGGCC__ACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU________ACAAAUGUACUCCGGUG (..(((.((((.((((.......((.....))...((((....))))...)))).)))).)))..)((....(((((..((((......))))..)))))....)). (-26.24 = -26.18 + -0.06)

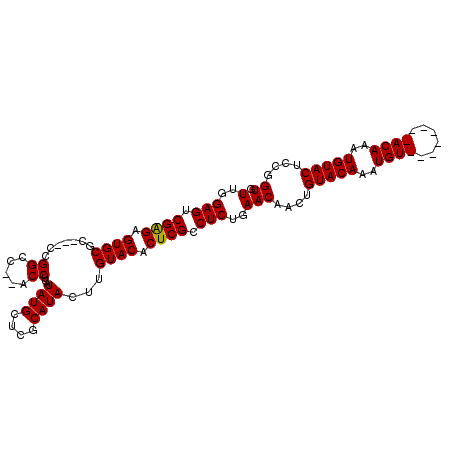
| Location | 13,344,743 – 13,344,845 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -27.99 |
| Energy contribution | -28.17 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13344743 102 - 22407834 CACCGGAGUACAUUUGUACAUUUGUACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGU--GGCCGG---GCGCACUCUCGACUCCAA ....((((((((..(((((....)))))..)))))....(((.(((((((..((..((.(((((....)))))....)--)..)).---.))).)))).)))))).. ( -36.00) >DroSec_CAF1 57028 96 - 1 CACCGGAGUACAAUUGU--------ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGUGAGGCCGG---GCGCACUCUCGACUCCAA ....(((((((((((((--------((....))))))))))..(((((((.((.(((..(((((....)))))...)))..))...---.))).))))..))))).. ( -33.90) >DroSim_CAF1 57369 96 - 1 CACCGGAGUACAUUUGU--------ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGUGAGGCCGC---GCGCACUCCCGGCUCCAA ....(((((...(((((--------(.(((((((((.((((......)))).))))))))).)))))).......((.(((((...---..)).))))).))))).. ( -34.70) >DroYak_CAF1 57602 105 - 1 CACUGGAGUACAUUUGUACAUUUCUACAUUUGUACAUUCGUUCAGAGGCGAGUGUACACGUAUGCGAGCAUAUGAGGU--GGCCAGGAGGCGCACUCUCGACUCCAA ...((((((.(...((((......))))..(((((((((((......))))))))))).(((((....)))))(((((--((((....))).))).)))))))))). ( -38.30) >consensus CACCGGAGUACAUUUGU________ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGU__GGCCGG___GCGCACUCUCGACUCCAA ....((((((((..((((......))))..)))))....(((.(((((((.........(((((....)))))..((.....))......))).)))).)))))).. (-27.99 = -28.17 + 0.19)

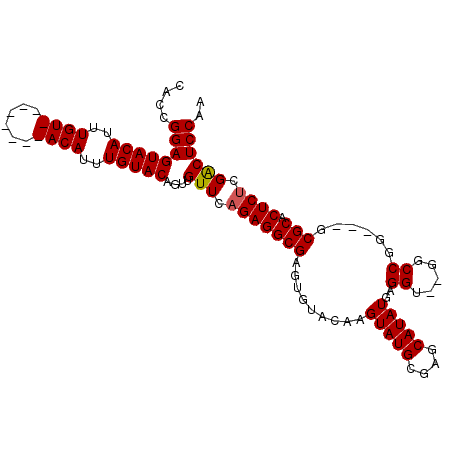
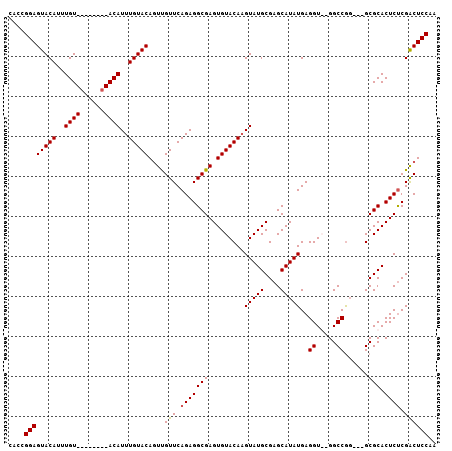
| Location | 13,344,767 – 13,344,873 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13344767 106 + 22407834 -ACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGUACAAAUGUACAAAUGUACUCCGGUGGACUACACU--GCUGCCACCUGGCGCCUCC -......((((....))))..........((((...........(((((..(((((....)))))..)))))...(((((.(......--...)))))).))))..... ( -25.60) >DroSec_CAF1 57053 101 + 1 CACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU--------ACAAUUGUACUCCGGUGGACUACACCGGGCUGCCACCUGGCGCCACC .......((((....))))...................((((.(((((....))--------))).))))....(((((.....)))))(((.(((....))))))... ( -27.90) >DroSim_CAF1 57394 101 + 1 CACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU--------ACAAAUGUACUCCGGUGGACUACACCGCGCUGCCACCUGGCGCCACC ..........((.(((......(((((.(((....))).....(((((....))--------)))..)))))...((((.....)))))))..(((....))))).... ( -23.70) >DroYak_CAF1 57629 106 + 1 -ACCUCAUAUGCUCGCAUACGUGUACACUCGCCUCUGAACGAAUGUACAAAUGUAGAAAUGUACAAAUGUACUCCAGUGGACUACUAC--GCUGCCACCUGGCGCCACC -.........((..((.....((((((.(((........))).))))))...((((....((((....))))(((...)))...))))--)).(((....))))).... ( -24.50) >consensus _ACCUCAUAUGCUCGCAUACUUGUACACUCGCCUCUGAACAACUGUACAAAUGU________ACAAAUGUACUCCGGUGGACUACACC__GCUGCCACCUGGCGCCACC .......((((....))))..........((((...........(((((..((((......))))..)))))...(((((.(...........)))))).))))..... (-19.88 = -20.25 + 0.38)


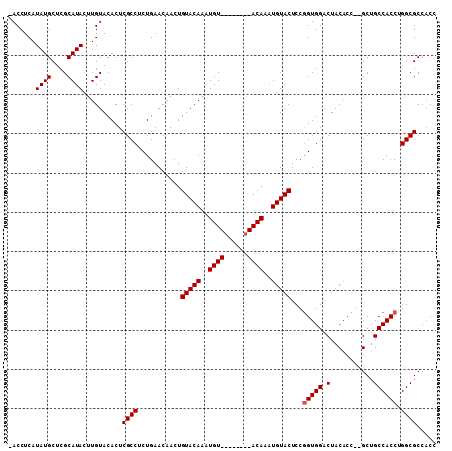
| Location | 13,344,767 – 13,344,873 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13344767 106 - 22407834 GGAGGCGCCAGGUGGCAGC--AGUGUAGUCCACCGGAGUACAUUUGUACAUUUGUACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGU- ....(((((....))).((--(..(((.(((...))).)))((((((((((((((...((((.((....)).)))).)))))))))))))).)))..)).........- ( -36.20) >DroSec_CAF1 57053 101 - 1 GGUGGCGCCAGGUGGCAGCCCGGUGUAGUCCACCGGAGUACAAUUGU--------ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGUG ((..(((((.(((....))).)))))...))(((...(((...((((--------(.(((((((((.((((......)))).))))))))).)))))...)))..))). ( -37.80) >DroSim_CAF1 57394 101 - 1 GGUGGCGCCAGGUGGCAGCGCGGUGUAGUCCACCGGAGUACAUUUGU--------ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGUG ....(((((....))).))((.(((((.(((...))).)))))((((--------(.(((((((((.((((......)))).))))))))).))))))).......... ( -35.30) >DroYak_CAF1 57629 106 - 1 GGUGGCGCCAGGUGGCAGC--GUAGUAGUCCACUGGAGUACAUUUGUACAUUUCUACAUUUGUACAUUCGUUCAGAGGCGAGUGUACACGUAUGCGAGCAUAUGAGGU- .((((..((((.(((..((--......))))))))).((((....))))....))))...(((((((((((......)))))))))))((((((....))))))....- ( -35.60) >consensus GGUGGCGCCAGGUGGCAGC__GGUGUAGUCCACCGGAGUACAUUUGU________ACAUUUGUACAGUUGUUCAGAGGCGAGUGUACAAGUAUGCGAGCAUAUGAGGU_ (((((((((....))).((.....)).).)))))...(((((..((((......))))..))))).....((((...((..(((((....)))))..))...))))... (-29.06 = -29.25 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:53 2006