
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,344,204 – 13,344,315 |
| Length | 111 |
| Max. P | 0.725807 |

| Location | 13,344,204 – 13,344,315 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -19.19 |
| Energy contribution | -18.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
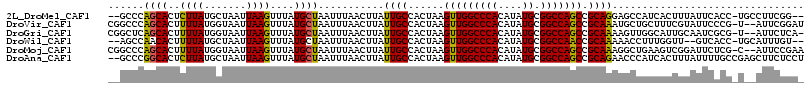
>2L_DroMel_CAF1 13344204 111 + 22407834 --CCGAAGGCA-GGUGAAUAAAGUGAUGGCUCCUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUAGCAUAAGAGUGCUGGGC-- --((.((((((-((.(.((......))..).))))).(((((((((....))))((((......))))...........))))).....)))...((((((....)))))))).-- ( -29.70) >DroVir_CAF1 78108 113 + 1 AUCCGAAU--A-CGGGAAUACGAAAGCAGCAUUUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUACCAUAAAAGUGCUGGGCCG .((((...--.-))))....(....)((((((((((((....))))...(((((((((......))))..((((((...........))))))....))))).))))))))..... ( -34.30) >DroGri_CAF1 178044 112 + 1 -UGAGAAU--A-CGCGAUUGCAAUGCCAACUUUUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUACCAUAAAAGUGCUGAGCCG -.......--.-(((((..(.........)..)))))...((((((((.(((((((((......))))..((((((...........))))))....)))))...)))).).))). ( -26.50) >DroWil_CAF1 114146 109 + 1 --ACAAAUGCA-GGUGAC--AACCAAAGGUUUUUGCGGUUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUAGCAUAAAAGUGUUGGCU-- --....((((.-(((...--.)))..((((((.(((((((((((.((....))))))))))...((((.....)))).....))).)))))).....)))).............-- ( -30.30) >DroMoj_CAF1 95855 113 + 1 UUCGGAAU--G-CGAGAAUCCGACUUCAGCCUUUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUACCAUAAAAGUGCUGGGCCG .(((((..--.-......))))).((((((((((((((....))))...(((((((((......))))..((((((...........))))))....))))))))).))))))... ( -34.10) >DroAna_CAF1 62818 114 + 1 AGGAGAAGCUCGGCAAAAUAAAGUGAUGGGUUCUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUAGCAUAAGAGUGCCGGGC-- .......((((((((..................(((((....)))))(((((..((((......))))..((((((...........))))))....)))))....))))))))-- ( -33.30) >consensus _UCCGAAU__A_CGAGAAUAAAAUGACAGCUUUUGCGGCUGGCCGCAUAUGUGGGCCAACUUAGUGGCAAUAAGUUAAAUUAGCAUAAACUUAAUUACCAUAAAAGUGCUGGGC__ ..........................((((...(((((....)))))..((((.((((......))))..((((((...........)))))).....)))).....))))..... (-19.19 = -18.92 + -0.28)


| Location | 13,344,204 – 13,344,315 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -16.27 |
| Energy contribution | -15.72 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13344204 111 - 22407834 --GCCCAGCACUCUUAUGCUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAGGAGCCAUCACUUUAUUCACC-UGCCUUCGG-- --....((((......))))...((((((...........))))))...((.....(((((((((....)).))))))).(((((.................))-)))....))-- ( -23.93) >DroVir_CAF1 78108 113 - 1 CGGCCCAGCACUUUUAUGGUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAAAUGCUGCUUUCGUAUUCCCG-U--AUUCGGAU .((((....((((...(((((((.(((((...........))))))))))))..)))).))))(((((.(((((....))))).))).))...........(((-.--...))).. ( -31.70) >DroGri_CAF1 178044 112 - 1 CGGCUCAGCACUUUUAUGGUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAAAAGUUGGCAUUGCAAUCGCG-U--AUUCUCA- .(((.((((...((..(((((((.(((((...........))))))))))))..)))))))))...(((((((((((((.......))))))........))))-)--)).....- ( -29.90) >DroWil_CAF1 114146 109 - 1 --AGCCAACACUUUUAUGCUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAACCGCAAAAACCUUUGGUU--GUCACC-UGCAUUUGU-- --.((((((........((....((((((...........))))))..))......))))))..(((.((((((.((((((..........)))))--)....)-))))).)))-- ( -24.74) >DroMoj_CAF1 95855 113 - 1 CGGCCCAGCACUUUUAUGGUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAAAGGCUGAAGUCGGAUUCUCG-C--AUUCCGAA (((((..((.......(((((((.(((((...........))))))))))))....(((((((((....)).))))))).))...)))))...(((((......-.--..))))). ( -36.80) >DroAna_CAF1 62818 114 - 1 --GCCCGGCACUCUUAUGCUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAGAACCCAUCACUUUAUUUUGCCGAGCUUCUCCU --((.(((((......(((....((((((...........))))))..........(((((((((....)).))))))).)))((.....)).........))))).))....... ( -27.30) >consensus __GCCCAGCACUUUUAUGCUAAUUAAGUUUAUGCUAAUUUAACUUAUUGCCACUAAGUUGGCCCACAUAUGCGGCCAGCCGCAAAAGCCGUCAUUGUAUUCACC_U__AUUCGGA_ ......((((..((((.......))))....))))...........((((......(((((((((....)).))))))).))))................................ (-16.27 = -15.72 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:50 2006