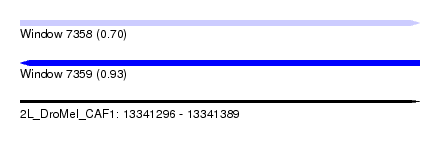
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,341,296 – 13,341,389 |
| Length | 93 |
| Max. P | 0.926348 |

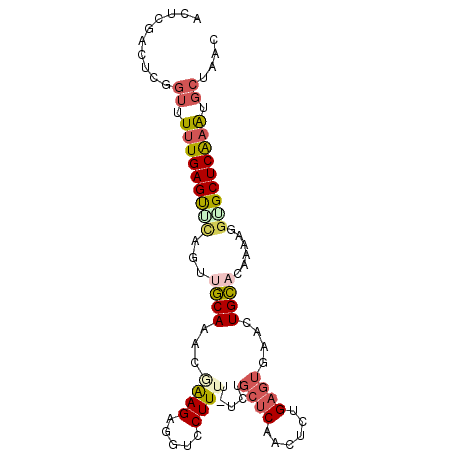
| Location | 13,341,296 – 13,341,389 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.72 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13341296 93 + 22407834 GUUAGCAUUCGAGCAUCUUUUGGACAGUUCACUCCGAGUUGAGCAGA-AAAAGGACCUCUUUGUUUGUAACUGUGCUCAAAAUCGGAGUCGAGU ......((((((((.((.....))..))))(((((((.(((((((.(-....((((......)))).....).)))))))..))))))).)))) ( -25.90) >DroSec_CAF1 53660 93 + 1 GUUAGCACUUGAGCACCUUAAGUGCAGUUCACUCAGUGUUGAGCAGA-AAAAGGAUCUCUUCGUUUGCAACUACACUCAAAAACAGAGUUGAGU ....(((((((((...)))))))))..((((((((((((...(((((-..(((.....)))..)))))....)))))........))).)))). ( -25.20) >DroSim_CAF1 53849 93 + 1 GUUAGCACUUGAGCACCUUUUGUGCAGUUCACUCAGUGUUGAGCAGA-AAAAGGACCUCUUCCUUUGCAACUAAACUCAAAAACAGAGUUGAGU .(((((((((((((((.....))))...)))...))))))))(((..-..(((((.....)))))))).(((.(((((.......))))).))) ( -23.50) >DroEre_CAF1 54810 90 + 1 GUUUGCAUUUGAGCACCUUUGCGGCAACU----CAGAGUUGAGUUGAAAAAAGGAGCUCUCAGCUUGCAACAGGACUCAAAGACCGAAUCGAGA ..(((((...((((.(((((....(((((----(......))))))...))))).))))......))))).....(((............))). ( -26.00) >DroYak_CAF1 53970 94 + 1 GUUUGCAUUUGAGCUCCUUUCGUGCAGCUCACUCUGAGUUGAGCUGUGAAAAGGAACUCUUAUUUUGCAACUGAACUCAGAAAGCGAGGCGAGU .(((((...((((((.(......).)))))).((((((((.((.((..((((((....))).)))..)).)).))))))))..)))))...... ( -32.40) >consensus GUUAGCAUUUGAGCACCUUUUGUGCAGUUCACUCAGAGUUGAGCAGA_AAAAGGACCUCUUAGUUUGCAACUGAACUCAAAAACAGAGUCGAGU (((.(((...(((..(((((....(.(((((........))))).)...)))))..)))......))))))...((((............)))) (-12.92 = -12.72 + -0.20)

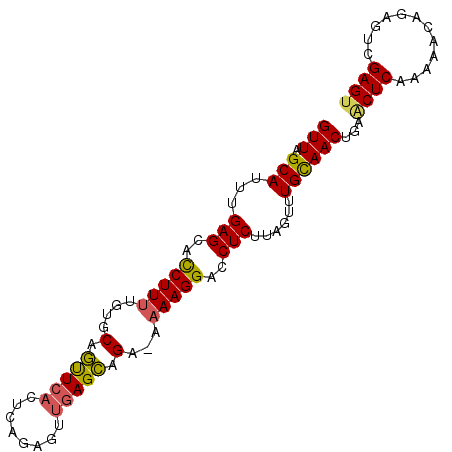
| Location | 13,341,296 – 13,341,389 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -10.34 |
| Energy contribution | -11.46 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13341296 93 - 22407834 ACUCGACUCCGAUUUUGAGCACAGUUACAAACAAAGAGGUCCUUUU-UCUGCUCAACUCGGAGUGAACUGUCCAAAAGAUGCUCGAAUGCUAAC ..((.(((((((..((((((...((.....))..(((((.....))-))))))))).)))))))))..............((......)).... ( -21.80) >DroSec_CAF1 53660 93 - 1 ACUCAACUCUGUUUUUGAGUGUAGUUGCAAACGAAGAGAUCCUUUU-UCUGCUCAACACUGAGUGAACUGCACUUAAGGUGCUCAAGUGCUAAC ..............((((((((((((........(((((.....))-)))(((((....))))).))))))))))))((..(....)..))... ( -25.20) >DroSim_CAF1 53849 93 - 1 ACUCAACUCUGUUUUUGAGUUUAGUUGCAAAGGAAGAGGUCCUUUU-UCUGCUCAACACUGAGUGAACUGCACAAAAGGUGCUCAAGUGCUAAC (((.(((((.......))))).)))((((.((((((((...)))))-)))(((((....)))))....)))).....((..(....)..))... ( -22.10) >DroEre_CAF1 54810 90 - 1 UCUCGAUUCGGUCUUUGAGUCCUGUUGCAAGCUGAGAGCUCCUUUUUUCAACUCAACUCUG----AGUUGCCGCAAAGGUGCUCAAAUGCAAAC ....((((((.....))))))...(((((......((((.(((((...(((((((....))----)))))....))))).))))...))))).. ( -26.80) >DroYak_CAF1 53970 94 - 1 ACUCGCCUCGCUUUCUGAGUUCAGUUGCAAAAUAAGAGUUCCUUUUCACAGCUCAACUCAGAGUGAGCUGCACGAAAGGAGCUCAAAUGCAAAC ....((...((...(((....)))..)).......(((((((((((..(((((((........)))))))...)))))))))))....)).... ( -33.10) >consensus ACUCGACUCGGUUUUUGAGUUCAGUUGCAAACGAAGAGGUCCUUUU_UCUGCUCAACUCUGAGUGAACUGCACAAAAGGUGCUCAAAUGCUAAC ..........((.(((((((((...((((...((((.....)))).....((((......))))....))))......))))))))).)).... (-10.34 = -11.46 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:48 2006