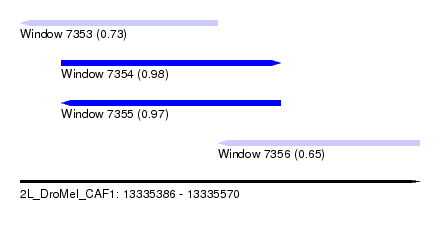
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,335,386 – 13,335,570 |
| Length | 184 |
| Max. P | 0.979564 |

| Location | 13,335,386 – 13,335,477 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -17.73 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
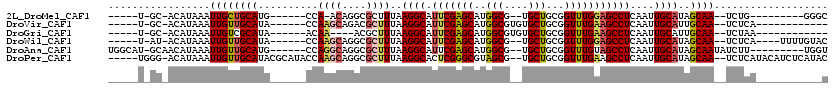
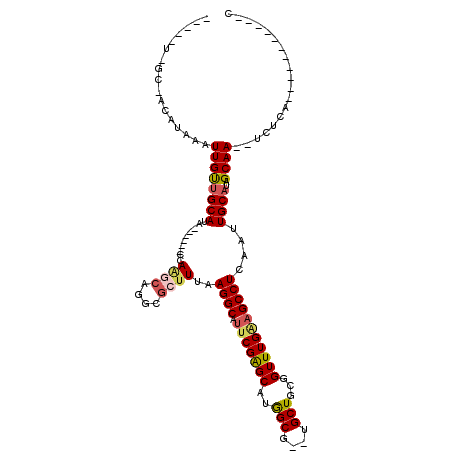
>2L_DroMel_CAF1 13335386 91 - 22407834 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAAAUGCACAAACAUAUGUA----- .(((((((.....((..((((((((..(((....)))...))))))))..))(((((((.......)-)))))).)))))))..........----- ( -23.70) >DroVir_CAF1 62814 90 - 1 UUUUGUGUAUGGCGUUUUGUUGAUUGAUGGUUAA-CAGAAAAUCAAUAUCACUAUGCUAAUGAGACA-AGCACGA--GCACGAGCACGAGCACG--- ..((((.(.((((((..((((((((..((.....-))...)))))))).....))))))...).)))-)((.((.--((....)).)).))...--- ( -22.10) >DroEre_CAF1 48848 91 - 1 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAAAUGCACAAACAUAUGUA----- .(((((((.....((..((((((((..(((....)))...))))))))..))(((((((.......)-)))))).)))))))..........----- ( -23.00) >DroWil_CAF1 84483 92 - 1 AUAUAUGUAUACCGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACUAGGCAUAAAUGCACAUACACAGACA----- .(((.(((((...((..((((((((..(((....)))...))))))))..))((((((..........)))))).))))).)))........----- ( -20.00) >DroYak_CAF1 48055 91 - 1 CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU-GGCAUAGAUGCACAAACAUAUGUG----- .((((((((((..((..((((((((..(((....)))...))))))))..))..(((((.......)-))))))))))))))..........----- ( -27.80) >DroAna_CAF1 48532 96 - 1 UUGUGUGUCCAACGUUUUGUUGAUUGACGGUUAGCCGGAAAAUCAAUAUCACUAUGCUAAUGAGACU-CACAUAAGUGCACAAACAUAUGUACACAG .((((.(((((.(((..((((((((..(((....)))...)))))))).....)))....)).))).-))))...(((.(((......))).))).. ( -23.90) >consensus CUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGACU_GGCAUAAAUGCACAAACAUAUGUA_____ ...(((((.....((..((((((((..(((....)))...))))))))..))((((((..........))))))...)))))............... (-17.73 = -17.82 + 0.09)


| Location | 13,335,405 – 13,335,506 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13335405 101 + 22407834 AUGCC-AG-------UCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGUUAGACACACAGGCCC---------CAGA--UUGCUAUGCAAUUGAGGCUCC .....-((-------(((((...((((((..((.((((((...(((....)))..))))))..................(....---------)..)--)..))))))...))))))).. ( -26.50) >DroVir_CAF1 62833 97 + 1 GUGCU-UG-------UCUCAUUAGCAUAGUGAUAUUGAUUUUCUG-UUAACCAUCAAUCAACAAAACGCCAUACACAAAA------------UGAGA--UUGCAAUGCAAUUGAGGCUUC .....-.(-------(((((...((((.(..((.((.(((((.((-(.........................))).))))------------).)))--)..).))))...))))))... ( -18.51) >DroGri_CAF1 165531 105 + 1 GUUCU-CGUUUUCAUUCUCAUUAGCAUUGCAAUAUUGAUUUCCUGGUUAACCAUCAAUCAACAAAACGACAGACACAAAA------------UUAGA--UUGCAAUGCAAUUGAGGCUUC .....-.........(((((...((((((((((.((((((...(((....)))..)))))).....(....)........------------....)--)))))))))...))))).... ( -23.30) >DroWil_CAF1 84502 107 + 1 AUGCCUAG-------UCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGGUAUACAUAUAUGUACAAAA----UGAGA--UUGCUAUGCAAUUGAGGCUCC ......((-------(((((...((((((..((.((((((...(((....)))..)))))).......(((((......)))))....----....)--)..))))))...))))))).. ( -29.50) >DroAna_CAF1 48556 103 + 1 AUGUG-AG-------UCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGCUAACCGUCAAUCAACAAAACGUUGGACACACAAACCA---------AAGAUAUUGCUAUGCAAUUGAGGCUAC .....-((-------(((((...((((((..(((((((((...(((....)))..)))).........((((.........)))---------).)))))..))))))...))))))).. ( -31.70) >DroPer_CAF1 62159 110 + 1 GUGCU-AG-------UCUCAUUAGCAUAGUGAUAUUGAUUUUCCGGUUAACCAUCAAUCAACAAAACGCCACUCACACAUGUAUGAGAUGUAUGAGA--UUGCUAUGCAAUUGAGGCUUC .....-((-------(((((...((((((..((.((((((....((....))...))))))..........((((.((((.......)))).)))))--)..))))))...))))))).. ( -32.10) >consensus AUGCU_AG_______UCUCAUUAGCAUAGUGAUAUUGAUUUUCUGGUUAACCAUCAAUCAACAAAACGCCAGACACACAAG___________UGAGA__UUGCUAUGCAAUUGAGGCUUC ...............(((((...(((((((((..((((((...(((....)))..))))))......(.....).........................)))))))))...))))).... (-17.62 = -17.40 + -0.22)

| Location | 13,335,405 – 13,335,506 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -19.15 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13335405 101 - 22407834 GGAGCCUCAAUUGCAUAGCAA--UCUG---------GGGCCUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGA-------CU-GGCAU ((.(((((((((((...))))--).))---------))))))...........((..((((((((..(((....)))...))))))))..)).((((((......-------.)-))))) ( -29.10) >DroVir_CAF1 62833 97 - 1 GAAGCCUCAAUUGCAUUGCAA--UCUCA------------UUUUGUGUAUGGCGUUUUGUUGAUUGAUGGUUAA-CAGAAAAUCAAUAUCACUAUGCUAAUGAGA-------CA-AGCAC ...........(((.(((...--(((((------------((....((((((.(...((((((((..((.....-))...)))))))).).)))))).)))))))-------))-)))). ( -25.00) >DroGri_CAF1 165531 105 - 1 GAAGCCUCAAUUGCAUUGCAA--UCUAA------------UUUUGUGUCUGUCGUUUUGUUGAUUGAUGGUUAACCAGGAAAUCAAUAUUGCAAUGCUAAUGAGAAUGAAAACG-AGAAC .....((((...(((((((((--.....------------......(.....)....((((((((..(((....)))...)))))))))))))))))...))))..........-..... ( -25.70) >DroWil_CAF1 84502 107 - 1 GGAGCCUCAAUUGCAUAGCAA--UCUCA----UUUUGUACAUAUAUGUAUACCGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGA-------CUAGGCAU ...((((...((((...))))--(((((----((.((((((....))))))..((..((((((((..(((....)))...))))))))..))......)))))))-------..)))).. ( -28.40) >DroAna_CAF1 48556 103 - 1 GUAGCCUCAAUUGCAUAGCAAUAUCUU---------UGGUUUGUGUGUCCAACGUUUUGUUGAUUGACGGUUAGCCGGAAAAUCAAUAUCACUAUGCUAAUGAGA-------CU-CACAU ((((.((((...((((((........(---------(((.........)))).....((((((((..(((....)))...))))))))...))))))...)))).-------))-.)).. ( -27.20) >DroPer_CAF1 62159 110 - 1 GAAGCCUCAAUUGCAUAGCAA--UCUCAUACAUCUCAUACAUGUGUGAGUGGCGUUUUGUUGAUUGAUGGUUAACCGGAAAAUCAAUAUCACUAUGCUAAUGAGA-------CU-AGCAC ..((.((((...(((((((..--.(((((((((.......)))))))))..))((..((((((((..(((....)))...))))))))..)))))))...)))).-------))-..... ( -35.20) >consensus GAAGCCUCAAUUGCAUAGCAA__UCUCA___________CUUGUGUGUCUAACGUUUUGUUGAUUGAUGGUUAACCAGAAAAUCAAUAUCACUAUGCUAAUGAGA_______CU_AGCAC .....((((...((((((...........................((.....))...((((((((..(((....)))...))))))))...))))))...))))................ (-19.15 = -18.90 + -0.25)



| Location | 13,335,477 – 13,335,570 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13335477 93 - 22407834 -----U-GC-ACAUAAAUUGCUGCAUG------CCA-ACAGGCGCUUUAAGGCAUUCGAGCAUGGCG--UGCUGCGGUUUGGAGCCUCAAUUGCAUAGCAA--UCUG---------GGGC -----.-..-...((((((((.(((((------(((-....(((((....)))......)).)))))--))).))))))))..(((((((((((...))))--).))---------)))) ( -34.90) >DroVir_CAF1 62904 93 - 1 -----U-GC-ACAUAAAUUGUUGCAUA------CCAAGCAGACGCUUUAAGGCAUUCGAGCAUGGCGUGUGCUGCGGUUUGAAGCCUCAAUUGCAUUGCAA--UCUCA------------ -----(-((-(....(((((..((..(------((.((((.(((((.....((......))..))))).))))..))).....))..)))))....)))).--.....------------ ( -28.90) >DroGri_CAF1 165610 89 - 1 -----U-GC-ACAUAAAUUGUCGCAUA------ACAA----ACGCUUUAAGGCAUUCGAGCAUGGCGUGUGCUGCGGUUUGAAGCCUCAAUUGCAUUGCAA--UCUAA------------ -----(-((-(....(((((..((...------.(((----(((((....)))...(((((((.....))))).)))))))..))..)))))....)))).--.....------------ ( -23.00) >DroWil_CAF1 84575 99 - 1 -----U-AU-ACAUAAAUUGUUGCAUA------CCAAGCAGGCGCUUUAAGGCAUUCGAGCAUGGCG--UGCUGCGGUUUGGAGCCUCAAUUGCAUAGCAA--UCUCA----UUUUGUAC -----.-.(-(((((((((((.(((..------(((.((.((.(((....)))..))..)).)))..--))).))))))))(((.....(((((...))))--)))).----...)))). ( -24.90) >DroAna_CAF1 48628 102 - 1 UGGCAU-GCAACAUAAAUUGUUGCAUG------CCAGGCAGGCGCUUUAAGGCAUUCGAGCAUGGCG--UGCUGCGGUUUGUAGCCUCAAUUGCAUAGCAAUAUCUU---------UGGU ((((((-((((((.....)))))))))------))).(((((((((.....((......))..))))--).))))(((.....)))...(((((...))))).....---------.... ( -39.50) >DroPer_CAF1 62231 110 - 1 -----UGGG-ACAUAAAUUGUUGCAUACGCAUACCAAGCAGGCGCUUUAAGGCACUCGGGCGUAGCG--UGCUGCGGUUUGAAGCCUCAAUUGCAUAGCAA--UCUCAUACAUCUCAUAC -----((((-(........(((((.((.(((......(.(((((((....)))..((((((((((..--..)))).)))))).)))))...))).))))))--)........)))))... ( -27.59) >consensus _____U_GC_ACAUAAAUUGUUGCAUA______CCAAGCAGGCGCUUUAAGGCAUUCGAGCAUGGCG__UGCUGCGGUUUGAAGCCUCAAUUGCAUAGCAA__UCUCA___________C .................((((((((..........((((....))))..((((.(((((((..(((....)))...)))))))))))....))))..))))................... (-17.88 = -18.35 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:45 2006