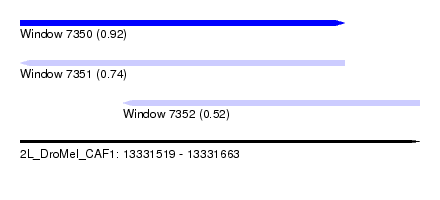
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,331,519 – 13,331,663 |
| Length | 144 |
| Max. P | 0.922029 |

| Location | 13,331,519 – 13,331,636 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13331519 117 + 22407834 ACAAAUGAUGGAUGAAUCAUUUGUCGGUGG--CGGGUAUUUUGCGCGAAUUGCCGAAAAUCGAUUCAGGGCGGCAUACCGAAAAGGAUGGAGACCUCAAUUGAUUUUGAGGUGUUUUGA .((((((((......))))))))(((((((--((.(.....).)))....(((((....((......)).))))))))))).......(((.(((((((......))))))).)))... ( -38.20) >DroPse_CAF1 57903 96 + 1 AUAAAUGAUGGAUGAAUCAUUAGUCAGAAAUGAAGGCCUGCCGUGCCAAAUGCUGCAAAUCGAUUAAAG-UUCCAUACAAAACGUGGUGAUGGCUUU---------------------- ...((((((......))))))..(((....)))((((((((.((.......)).)))...........(-((((((.......)))).)))))))).---------------------- ( -17.40) >DroSec_CAF1 44113 117 + 1 ACAAAUGAUGGAUGAAUCAUUUGUCGGUGG--CGGGUAUUUCGCGCGAAUUGCCGAAAAUCGAUUCGGGGCGGCAUUCCGAAAAGGAUGGAGACCUCAAUUGAUUUUGAGGUGUUUUGA .((((((((......))))))))((((..(--((.(.....).)))....(((((....(((...)))..)))))..)))).......(((.(((((((......))))))).)))... ( -36.40) >DroSim_CAF1 43418 117 + 1 ACAAAUGAUGGAUGAAUCAUUUGUCGGUGG--CGGGUAUUUCGCGCGAAUUGCCGAAAAUCGAUUCAGGGCGGCAUACCGAAAAGGAUGGAGACCUCAAUUGAUUUUGAGGUGUUUUGA .((((((((......))))))))(((((((--((.(.....).)))....(((((....((......)).))))))))))).......(((.(((((((......))))))).)))... ( -39.30) >DroEre_CAF1 45003 117 + 1 ACAAAUGAUGGAUGAAUCAUUUGUCGGUGG--CGGGUAUUUCGCGCGAAUUGCCGAAAAUCGAUUCAGGGCGGCAUACUGAAAUGGAUAGAGACCUCAAUUGAUUUUGAGGUGUUUCAA .((((((((......))))))))(((((((--((.(.....).)))....(((((....((......)).))))))))))).......(((.(((((((......))))))).)))... ( -37.20) >DroYak_CAF1 44045 117 + 1 ACAAGUGAUGGAUGAAUCAUUUGUCGGUGG--CGGGUAUUUUGUGCGAAUUGCCGAAAAUCGAUUCAGGGCGGCAUACUGAAAGGGAUAGAGACCUCAAUUGAUUUUGAGGUGAUUCAA (((((((((......))))))))).....(--((.(.....).)))((((..((.((((((((((.(((........((....))........))).))))))))))..))..)))).. ( -35.89) >consensus ACAAAUGAUGGAUGAAUCAUUUGUCGGUGG__CGGGUAUUUCGCGCGAAUUGCCGAAAAUCGAUUCAGGGCGGCAUACCGAAAAGGAUGGAGACCUCAAUUGAUUUUGAGGUGUUUUGA .((((((((......))))))))((((..(..((.((.....)).))..)..))))...((.((((....(((....)))....)))).)).(((((((......)))))))....... (-24.90 = -25.10 + 0.20)

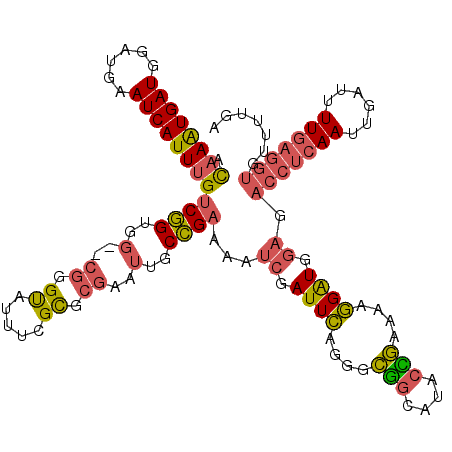
| Location | 13,331,519 – 13,331,636 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -13.79 |
| Energy contribution | -15.43 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13331519 117 - 22407834 UCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCAAAAUACCCG--CCACCGACAAAUGAUUCAUCCAUCAUUUGU .......(((((((......)))))))...........(((((.(((((....(((.....))))))))....(((.........))--).)))))((((((((......)))))))). ( -27.20) >DroPse_CAF1 57903 96 - 1 ----------------------AAAGCCAUCACCACGUUUUGUAUGGAA-CUUUAAUCGAUUUGCAGCAUUUGGCACGGCAGGCCUUCAUUUCUGACUAAUGAUUCAUCCAUCAUUUAU ----------------------...(((....(((.((.((((((.((.-......)).))..)))).)).)))...))).((...(((((.......))))).....))......... ( -12.90) >DroSec_CAF1 44113 117 - 1 UCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGAAUGCCGCCCCGAAUCGAUUUUCGGCAAUUCGCGCGAAAUACCCG--CCACCGACAAAUGAUUCAUCCAUCAUUUGU .......(((((((......)))))))...........((((...((....(((((......)))))......))(((.......))--)..))))((((((((......)))))))). ( -28.30) >DroSim_CAF1 43418 117 - 1 UCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAAAUACCCG--CCACCGACAAAUGAUUCAUCCAUCAUUUGU .......(((((((......)))))))...........(((((.(((((....(((.....))))))))....(.(((.......))--)))))))((((((((......)))))))). ( -28.90) >DroEre_CAF1 45003 117 - 1 UUGAAACACCUCAAAAUCAAUUGAGGUCUCUAUCCAUUUCAGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAAAUACCCG--CCACCGACAAAUGAUUCAUCCAUCAUUUGU ((((...(((((((......)))))))..........(((((.........))))))))).(((((((.....)).)))))......--......(((((((((......))))))))) ( -24.50) >DroYak_CAF1 44045 117 - 1 UUGAAUCACCUCAAAAUCAAUUGAGGUCUCUAUCCCUUUCAGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCACAAAAUACCCG--CCACCGACAAAUGAUUCAUCCAUCACUUGU .(((((((.(((((......)))))(((...(((...(((((.........)))))..)))....(((..................)--))...)))...)))))))............ ( -20.37) >consensus UCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAAAUACCCG__CCACCGACAAAUGAUUCAUCCAUCAUUUGU .......(((((((......))))))).................(((((....(((.....))))))))..........................(((((((((......))))))))) (-13.79 = -15.43 + 1.64)
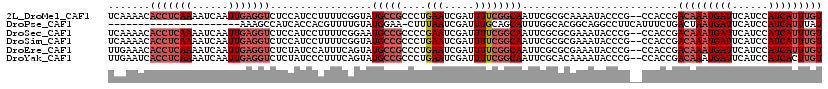
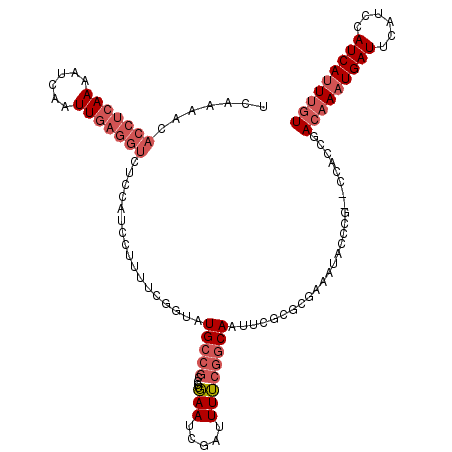
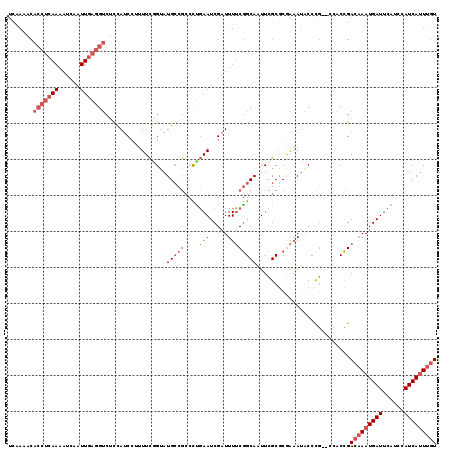
| Location | 13,331,556 – 13,331,663 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.16 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13331556 107 - 22407834 GAAUUGGAUUACAAAAUCUCUUGUUCUUCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCAAA ((((((((..((((......))))...))......((..((((((.(((.(((.(..(((((.....)).)))..).))).))).)))))).))))))))....... ( -21.80) >DroSec_CAF1 44150 107 - 1 GAAUUGGAUUACAAAAUCUCCCGUUCUUCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGAAUGCCGCCCCGAAUCGAUUUUCGGCAAUUCGCGCGAA ((((((((((....))))..(((....((.....(((((((......)))))))..........(((((.........)))))..))....)))))))))....... ( -23.30) >DroSim_CAF1 43455 107 - 1 GAAUUGGAUUAUAAAAUCUCCCGUUCUUCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAA ((((((((((....))))..(((....((.....(((((((......)))))))............(((....))).........))....)))))))))....... ( -21.70) >DroEre_CAF1 45040 105 - 1 GAAUUGA-UUAUUGAAACUGUCGAU-UUUGAAACACCUCAAAAUCAAUUGAGGUCUCUAUCCAUUUCAGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAA ((((((.-.((((((((.((..(((-...((...(((((((......))))))).)).))))))))))))).......(((((......)))))))))))....... ( -23.20) >DroYak_CAF1 44082 107 - 1 GAAUUGAAUUAUUUAAUCUGUGGUUUUUUGAAUCACCUCAAAAUCAAUUGAGGUCUCUAUCCCUUUCAGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCACAAA ((((((.............(((((...(((((..(((((((......)))))))..........)))))...))))).(((((......)))))))))))....... ( -25.00) >consensus GAAUUGGAUUAUAAAAUCUCUCGUUCUUCAAAACACCUCAAAAUCAAUUGAGGUCUCCAUCCUUUUCGGUAUGCCGCCCUGAAUCGAUUUUCGGCAAUUCGCGCGAA ((((((............................(((((((......)))))))........................(((((......)))))))))))....... (-17.52 = -17.36 + -0.16)

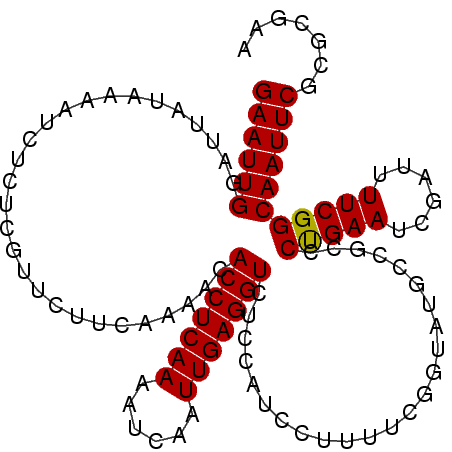
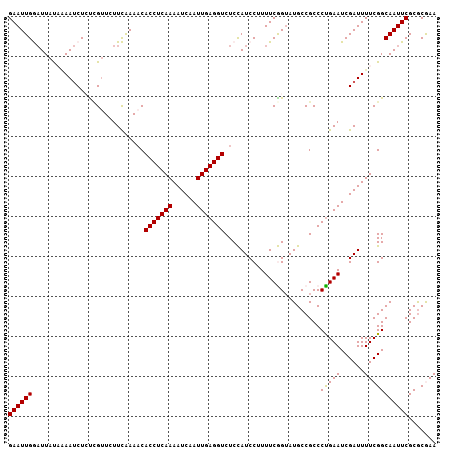
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:41 2006