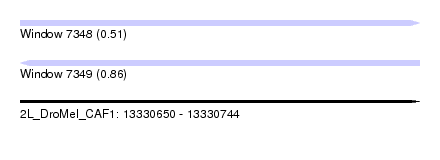
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,330,650 – 13,330,744 |
| Length | 94 |
| Max. P | 0.858675 |

| Location | 13,330,650 – 13,330,744 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13330650 94 + 22407834 UUUGAGGCAGCCAAUGUCAGAGAACGCCAUUCGCUGAUUGGCUGCGCCCGCUGCAUGCUGCAAUCGGAAUUGUUUUUCGCCUGGCCUGGCUU---GG ...((((((((((..(((((.(((.....))).))))))))))))..(((.(((.....)))..)))........)))(((......)))..---.. ( -31.40) >DroPse_CAF1 56841 92 + 1 UUUGUGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGC--CUGCAGCAUGCUGCAGGACCAAUUGUUGUU-GCC--ACUUCGCUUCCCGU ...(((((((((((((((((.((.......)).)))))(((...(--((((((....))))))).)))))))..)))-)))--))............ ( -35.70) >DroSec_CAF1 43252 94 + 1 UUUGAGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGCGCCCGCUGCAUGCAGCAAUCGGAAUUGUUUUUUGCCUGGCCUGGCUU---GG (((((.(((((((..(((((.((.......)).))))))))))))....((((....))))..)))))..........(((......)))..---.. ( -32.20) >DroEre_CAF1 44139 89 + 1 UUUGAGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGCGCCCGCUGCAUGCUGCAAUCGGAAUUGUUUCUCGCCUGGCCU--------GG ...((((((((((..(((((.((.......)).))))))))))))..(((.(((.....)))..)))........))).((......--------)) ( -28.30) >DroAna_CAF1 44267 77 + 1 UUUGUGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGC-CGCGCUGCAUGCUGCAAGCCCAAUUGUUUUU-------------------G ...((((((((((..(((((.((.......)).))))))))))))-)))(((((.....)).)))............-------------------. ( -31.10) >DroPer_CAF1 56338 92 + 1 UUUGUGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGC--CUGCAGCAUGCUGCAUGACCAAUUGUUGUU-GCC--ACUUCGCUUCCCGU ...((((((((((((((((......((((.((...)).))))...--.(((((....)))))))))..))))..)))-)))--))............ ( -30.10) >consensus UUUGAGGCAGCCAAUGUCAGAGAACGCCACUCGCUGAUUGGCUGC_CCCGCUGCAUGCUGCAAGCCCAAUUGUUUUU_GCC__GCCU_GCUU___GG ......(((((((..(((((.((.......)).))))))))))))....((.(....).)).................................... (-20.22 = -20.22 + -0.00)

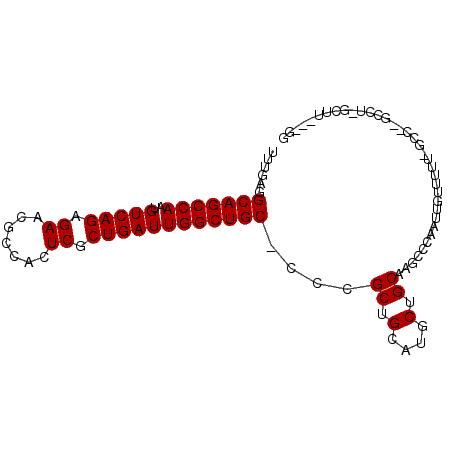
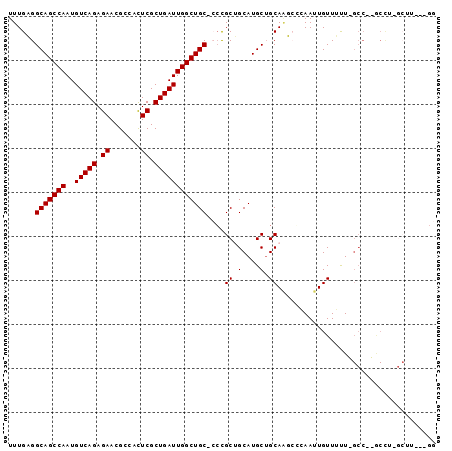
| Location | 13,330,650 – 13,330,744 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13330650 94 - 22407834 CC---AAGCCAGGCCAGGCGAAAAACAAUUCCGAUUGCAGCAUGCAGCGGGCGCAGCCAAUCAGCGAAUGGCGUUCUCUGACAUUGGCUGCCUCAAA ..---..(((......)))((........((((.((((.....)))))))).((((((((((((.((((...)))).))))..)))))))).))... ( -33.00) >DroPse_CAF1 56841 92 - 1 ACGGGAAGCGAAGU--GGC-AACAACAAUUGGUCCUGCAGCAUGCUGCAG--GCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCACAAA ..((((..(((..(--(..-..))....))).))))((((....)))).(--((((((((((((.(((.....))).))))..)))))))))..... ( -36.70) >DroSec_CAF1 43252 94 - 1 CC---AAGCCAGGCCAGGCAAAAAACAAUUCCGAUUGCUGCAUGCAGCGGGCGCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCUCAAA ..---..(((......))).................((((....))))(((.((((((((((((.(((.....))).))))..)))))))))))... ( -33.80) >DroEre_CAF1 44139 89 - 1 CC--------AGGCCAGGCGAGAAACAAUUCCGAUUGCAGCAUGCAGCGGGCGCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCUCAAA ..--------.........(((.......((((.((((.....)))))))).((((((((((((.(((.....))).))))..)))))))))))... ( -30.70) >DroAna_CAF1 44267 77 - 1 C-------------------AAAAACAAUUGGGCUUGCAGCAUGCAGCGCG-GCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCACAAA .-------------------..........(.(((.((.....))))).)(-((((((((((((.(((.....))).))))..)))))))))..... ( -30.00) >DroPer_CAF1 56338 92 - 1 ACGGGAAGCGAAGU--GGC-AACAACAAUUGGUCAUGCAGCAUGCUGCAG--GCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCACAAA ....((..(((..(--(..-..))....))).)).(((((....)))))(--((((((((((((.(((.....))).))))..)))))))))..... ( -33.50) >consensus CC___AAGC_AGGC__GGC_AAAAACAAUUCCGAUUGCAGCAUGCAGCGGG_GCAGCCAAUCAGCGAGUGGCGUUCUCUGACAUUGGCUGCCACAAA ..................................((((.(....).))))..((((((((((((.(((.....))).))))..))))))))...... (-20.91 = -20.80 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:39 2006