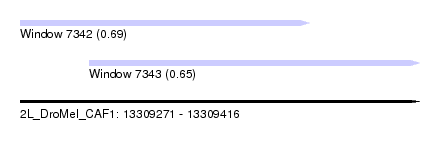
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,309,271 – 13,309,416 |
| Length | 145 |
| Max. P | 0.688377 |

| Location | 13,309,271 – 13,309,376 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -27.51 |
| Energy contribution | -26.18 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
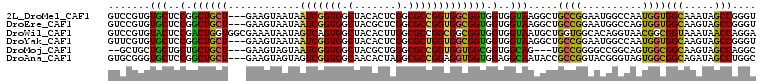
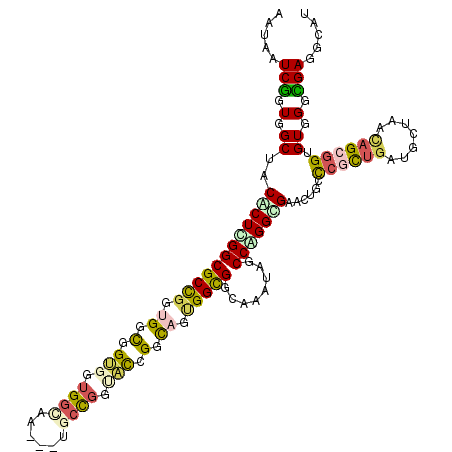
>2L_DroMel_CAF1 13309271 105 + 22407834 GUCCGUGUGCUCCGGCUGCU---GAAGUAAUAAUCGGUGGCUACACUCGGCGCCGGUGGCGGUGGUGGUAAGGCUGCCGGAAUGGCCAAUGGUGGCAAAUAGCCGGGU ..........((((((((..---..........((((..(((.(((.((.((((...)))).)))))....)))..))))....((((....))))...)))))))). ( -42.50) >DroEre_CAF1 23471 105 + 1 GUCCGUGUGCUCCGGCUGCU---GAAGUAAUAAUCGGUGGCUACGCUCGGCGCCGGUGGCGGUGGUGGUAAGGCUGCCGGAAUGGCCAGUGGUGGCAAGUAGCCGGGU ..........((((((((((---..........((((..(((..((.((.(((((....))))).))))..)))..))))....((((....)))).)))))))))). ( -47.50) >DroWil_CAF1 29042 108 + 1 GUCCGUGUACUCCGACUGGUGGCGAAAUAAUAGUCAGUGGCUACACUUGGCGCCGGCGGCGGUGGUGGUAAUGCUGGUGGCACAGGUAACGGCGGUAAAUAACCAGGA .(((.......(((.(((.(((((......((((.....)))).......))))).))))))((((..((.(((((...((....))..))))).))....))))))) ( -34.12) >DroYak_CAF1 22042 105 + 1 GUUCGUGUGCUCCGGCUGCU---GAAGUAAUAAUCGGUGGCUACACUCGGCGCUGGUGGCGGUGGUGGUAAGGCUGCCGGAAUGGCCAAUGGUGGCAAGUAGCCGGGU ..........((((((((((---..........((((..(((.(((.((.(((.....))).)))))....)))..))))....((((....)))).)))))))))). ( -45.90) >DroMoj_CAF1 30437 100 + 1 --GCUGCUGCUGCUGCUGCU---GAAGUAGUAAUCGGUGGCUACGCUGGGCGCCGGUGGUGGCGGUGGCAG---UGCCGGGGCCGGCAGUGGCGGCAAGUAGCCAGGC --(((((((((((..(((((---...........(((((....)))))(((.((((((.((.(...).)).---)))))).))))))))..))))).))))))..... ( -56.30) >DroAna_CAF1 24751 105 + 1 GUGCGGGUGCUCCGGCUGCU---GAAGUAGUAGUCGGUGGCAACACUAGGCGCCGGAGGUGGUGGAGGCAAUACCGCCGGUACGGGUAGUGGCGGCAGAUAGCCUGGC ...(((((.(((((((((((---(...)))))(((((((....)))).))))))))))..((((.......))))((((.(((.....))).)))).....))))).. ( -46.70) >consensus GUCCGUGUGCUCCGGCUGCU___GAAGUAAUAAUCGGUGGCUACACUCGGCGCCGGUGGCGGUGGUGGUAAGGCUGCCGGAACGGCCAAUGGCGGCAAAUAGCCGGGU .......(((..(.((((((............(((((((.(.......).))))))))))))).)..))).....((((..........))))(((.....))).... (-27.51 = -26.18 + -1.33)
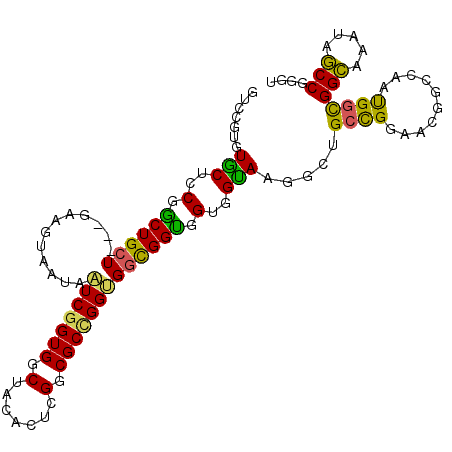
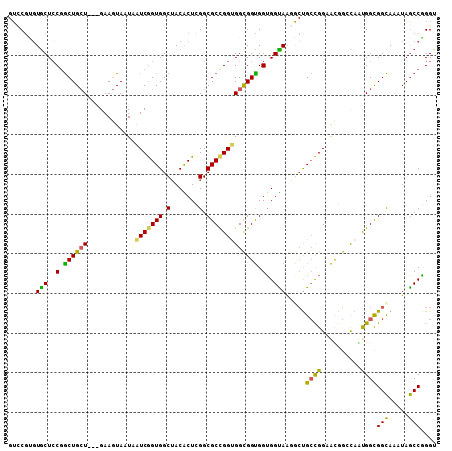
| Location | 13,309,296 – 13,309,416 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Mean single sequence MFE | -52.65 |
| Consensus MFE | -30.81 |
| Energy contribution | -31.02 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13309296 120 + 22407834 AAUAAUCGGUGGCUACACUCGGCGCCGGUGGCGGUGGUGGUAAGGCUGCCGGAAUGGCCAAUGGUGGCAAAUAGCCGGGUGAAGUUCCCGCUGAUGCUAACAGCGGUGUCGGCGAGGCAU .....((((..(((.((((((((.((((..((..((....))..))..))))....((((....)))).....)))))))).)))..))(((((((((......)))))))))))..... ( -54.80) >DroVir_CAF1 26322 117 + 1 AAUAAUCGGUGGCUACGCUCGGCGCUGGUGGCGGUGGUGGCAG---CACCGGUUCCGGCAGAGGCGGCAGAUAGCCGGGUGAGCUGCCCGCUGAUGCUAAUAGCGGUGUGGGCGACGCGU ........((((((.((((((((.(((.((.(..((.(((.((---(....)))))).))..).)).)))...)))))))))))(((((((...(((.....)))..))))))).))).. ( -53.30) >DroGri_CAF1 26635 117 + 1 AGUAGUCGGUGGCUACGCUUGGCGCCGGUGGUGGCGGUGGCAG---UUCCGGUGCCGGCAGUGGUGGCAGAUAACCCGGCGAACUGCUGUCCGAUCCCAACAGCGGUGUGGGUGAGGCGU .(((((.....)))))(((((.(((((.((.(((((.(((...---..))).))))).)).))))).).....(((((....((((((((.........)))))))).))))).)))).. ( -52.00) >DroWil_CAF1 29070 120 + 1 AAUAGUCAGUGGCUACACUUGGCGCCGGCGGCGGUGGUGGUAAUGCUGGUGGCACAGGUAACGGCGGUAAAUAACCAGGAGAACUUCCCGCUGAAGCUAAGAGCGGUGUGGGUGAGGCAU ...........(((.(((.(.(((((..((((((((((..((.(((((...((....))..))))).))....))))(((....)))))))))..((.....))))))).)))).))).. ( -38.50) >DroMoj_CAF1 30460 117 + 1 AGUAAUCGGUGGCUACGCUGGGCGCCGGUGGUGGCGGUGGCAG---UGCCGGGGCCGGCAGUGGCGGCAAGUAGCCAGGCGAGCUGCCCGUUGACGAGAGCAGCGGUGUGGGCGACGUCU .........(((((((....(.(((((.((.((((..((((..---.))))..)))).)).))))).)..)))))))((((.((..(((((((.(....))))))).)..).)..)))). ( -58.80) >DroAna_CAF1 24776 120 + 1 AGUAGUCGGUGGCAACACUAGGCGCCGGAGGUGGUGGAGGCAAUACCGCCGGUACGGGUAGUGGCGGCAGAUAGCCUGGCGAGCUGCCCGCUGAGGCUAGCAGGGGCGUCGGCGAGGCAU ....(((((((....)))).)))(((...(((((((.......))))))).....(((((((..((.(((.....))).)).)))))))(((((.(((......))).)))))..))).. ( -58.50) >consensus AAUAAUCGGUGGCUACACUCGGCGCCGGUGGCGGUGGUGGCAA___UGCCGGUACCGGCAGUGGCGGCAAAUAGCCAGGCGAACUGCCCGCUGAUGCUAACAGCGGUGUGGGCGAGGCAU .....(((.(.((..((((((((((((.((.(.(((.((((......)))).))).).)).)))))........)))))))......((((((.......)))))).)).).)))..... (-30.81 = -31.02 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:33 2006