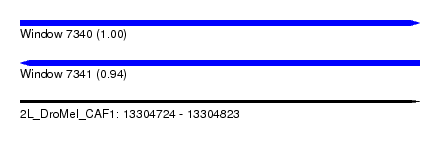
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,304,724 – 13,304,823 |
| Length | 99 |
| Max. P | 0.996543 |

| Location | 13,304,724 – 13,304,823 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13304724 99 + 22407834 CUAAAA-ACUAACCACUGUGCUCAUGCC-----------------AAAUGGCGCUCCUCCUCGCAGAUGGCACUGCGCGUCGCCUCGCACUGGGCACAGUGCUUCUCCAGAAACUUC ......-......((((((((((((((.-----------------....((((......(.(((((......))))).).))))..))).)))))))))))................ ( -32.50) >DroVir_CAF1 21155 112 + 1 CUUAA--ACUAACCACUGUGCGCGUGCGAAUCG--GCCGC-CGCCAGGUGGCGCUGCUCCUCGUUGACGGCUUGACGCGCGGCCUCGCAUUGCGCACAGUGCUUCUCCAGAAACUUC .....--......((((((((((((((((...(--(((((-((((....)))((((.((......)))))).....).))))))))))).)))))))))))................ ( -50.50) >DroPse_CAF1 22611 115 + 1 CUAAA--GCUAACCACUGUGCCCGCCCCCGCCCCCGCCAUCAGCGAGGUGGCGCUGCUCCUCGCUGAUGGCGUUGCGCGUCGCCUCGCACUGGGCACAGUGCUUCUCCAAAAACUUC .....--......((((((((((......((...((((((((((((((.(((...)))))))))))))))))..))(((......)))...))))))))))................ ( -57.60) >DroGri_CAF1 21033 112 + 1 CUUAA--ACUAACCACUGUGCGCGUGAGAUUCU--GCCGU-CGCCAGAUGGCGCUGCUCUUCGCUGACCGCGAGACGCGCCGCCUCACAUUGCGCACAGUGCUUCUCCAGAAACUUC .....--......(((((((((((((((.....--(((((-(....))))))((.(((((.(((.....)))))).))))...)))))...))))))))))................ ( -44.80) >DroMoj_CAF1 24168 112 + 1 CUUAA--ACUAACCACUGUGCGCAUGCGUUUCG--GCCGC-CGCCAGAUGGCGCUGCUCCUCGCUGACAGCCUGUCGUGCCGCCUCGCAUUGCGCACAGUGCUUCUCCAGAAACUUC .....--......(((((((((((((((...((--((((.-((((....)))((((.((......))))))..).)).))))...)))).)))))))))))................ ( -46.20) >DroAna_CAF1 18473 103 + 1 CUAAAACUCUAACCACUGGACUCAGGCGCG--------------GAGGUGGCGCUGCUCCUCGCUAAUGGCGCUGCGCGUCGCCUCGCACUGGGCACAGUGCUUCUCCAGAAACUUC .............(((((..(((((..(((--------------.(((((((((.((....(((.....)))..)))))))))))))).)))))..)))))................ ( -42.40) >consensus CUAAA__ACUAACCACUGUGCGCAUGCGAGUC___GCCG__CGCCAGAUGGCGCUGCUCCUCGCUGACGGCGUGGCGCGCCGCCUCGCACUGCGCACAGUGCUUCUCCAGAAACUUC .............((((((((((((((........((.....))..((.((((.(((.....((.....)).....))).))))))))).)))))))))))................ (-26.45 = -26.82 + 0.36)

| Location | 13,304,724 – 13,304,823 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -47.33 |
| Consensus MFE | -26.20 |
| Energy contribution | -28.23 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13304724 99 - 22407834 GAAGUUUCUGGAGAAGCACUGUGCCCAGUGCGAGGCGACGCGCAGUGCCAUCUGCGAGGAGGAGCGCCAUUU-----------------GGCAUGAGCACAGUGGUUAGU-UUUUAG .......((((((((.((((((((...((((.(((((.(.(((((......)))))....)...))))...)-----------------.))))..)))))))).)...)-)))))) ( -35.80) >DroVir_CAF1 21155 112 - 1 GAAGUUUCUGGAGAAGCACUGUGCGCAAUGCGAGGCCGCGCGUCAAGCCGUCAACGAGGAGCAGCGCCACCUGGCG-GCGGC--CGAUUCGCACGCGCACAGUGGUUAGU--UUAAG ...(((((....)))))(((((((((..(((((((((((((.((....((....))..))))..((((....))))-)))))--)...))))).))))))))).......--..... ( -50.90) >DroPse_CAF1 22611 115 - 1 GAAGUUUUUGGAGAAGCACUGUGCCCAGUGCGAGGCGACGCGCAACGCCAUCAGCGAGGAGCAGCGCCACCUCGCUGAUGGCGGGGGCGGGGGCGGGCACAGUGGUUAGC--UUUAG .........((..((.((((((((((.(((((......)))))..((((((((((((((.((...))..))))))))))))))...((....)))))))))))).))..)--).... ( -62.20) >DroGri_CAF1 21033 112 - 1 GAAGUUUCUGGAGAAGCACUGUGCGCAAUGUGAGGCGGCGCGUCUCGCGGUCAGCGAAGAGCAGCGCCAUCUGGCG-ACGGC--AGAAUCUCACGCGCACAGUGGUUAGU--UUAAG ...(((((....)))))(((((((((...((((((.(((((.((((((.....))).)))...))))).((((.(.-..).)--))).))))))))))))))).......--..... ( -48.90) >DroMoj_CAF1 24168 112 - 1 GAAGUUUCUGGAGAAGCACUGUGCGCAAUGCGAGGCGGCACGACAGGCUGUCAGCGAGGAGCAGCGCCAUCUGGCG-GCGGC--CGAAACGCAUGCGCACAGUGGUUAGU--UUAAG ...(((((....)))))((((((((((.((((...((((.((.(..((((((......)).))))(((....))))-.))))--))...)))))))))))))).......--..... ( -50.20) >DroAna_CAF1 18473 103 - 1 GAAGUUUCUGGAGAAGCACUGUGCCCAGUGCGAGGCGACGCGCAGCGCCAUUAGCGAGGAGCAGCGCCACCUC--------------CGCGCCUGAGUCCAGUGGUUAGAGUUUUAG .(((.((((((.....(((((..(.((((((((((.(..((((.((.((........)).)).))))).))).--------------)))).))).)..))))).)))))).))).. ( -36.00) >consensus GAAGUUUCUGGAGAAGCACUGUGCCCAAUGCGAGGCGACGCGCAACGCCAUCAGCGAGGAGCAGCGCCACCUGGCG__CGGC___GAAGCGCACGCGCACAGUGGUUAGU__UUAAG ...(((((....)))))(((((((((.(((((.((((..((....(((.....)))....))..)))).....((.....)).......)))))))))))))).............. (-26.20 = -28.23 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:31 2006