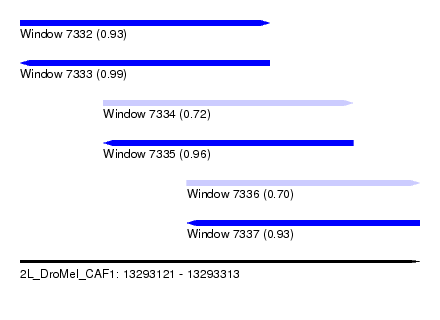
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,293,121 – 13,293,313 |
| Length | 192 |
| Max. P | 0.985463 |

| Location | 13,293,121 – 13,293,241 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -32.68 |
| Energy contribution | -33.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293121 120 + 22407834 CCAAGAAGGACUCGAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAACAGAAGCAGAAGGAGCAGCAGAAGGCUAUGGAGGCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUG .((((((...((((...........))))((((..(((((((((.....(..((.......))..).)))))))))(((((((.((......))))))))).........)))))))))) ( -40.80) >DroSec_CAF1 5990 120 + 1 CAAAGAAGGACUCGAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCG .......................((.(((((((.....((((((..((....))....(((((........)))))))))))(((...(((....)))))).........))))))))). ( -38.80) >DroSim_CAF1 5975 120 + 1 CCAAGAAGGACUCAAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAGCAAAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCG ((.....))..............((.(((((((.....((((((..((....))....(((((........)))))))))))(((...(((....)))))).........))))))))). ( -39.10) >DroEre_CAF1 7015 120 + 1 CGAAGAAGGACUCGAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAGCAGAAGCAGAAGGAGCAGAAGAAGGCUCUGGAGGCAGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUGUUG ..((((.((.(((......((.....))(((((.(((.((((((..((....))....(((((........))))))))))).((....))..)))))))).....))).)))))).... ( -39.10) >DroYak_CAF1 6148 120 + 1 CAAAGAAGGACUCGAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCAGCCAAGGCUGGUGCCUCCAAGAAGGGACCACUUGUUG ..........((((...........))))...((((((((((((..((....))....(((((........)))))(((((((.(((....))))))))))..)))))..)))..)))). ( -38.30) >consensus CAAAGAAGGACUCGAAGAAUCUAGACGAGGAGGACAUGGCCUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUG ..((((.((.(((......((.....))(((((.(((.((((((..((....))....(((((........))))))))))).((....))..)))))))).....))).)))))).... (-32.68 = -33.68 + 1.00)

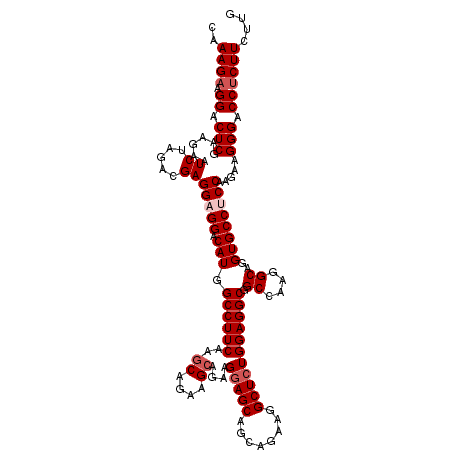

| Location | 13,293,121 – 13,293,241 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293121 120 - 22407834 CAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGCCUCCAUAGCCUUCUGCUGCUCCUUCUGCUUCUGUUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUCGAGUCCUUCUUGG ((((((((((.(......(((((((.((......))..)))))))..((((((.((.((.......))....))..))))))...).))))((((...........))))...)))))). ( -40.20) >DroSec_CAF1 5990 120 - 1 CGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUCGAGUCCUUCUUUG ((((..((((.(...(((.((((((..........))))))...)))((((((.((.((.......))....))..))))))...).))))))))....(((((...)))))........ ( -37.40) >DroSim_CAF1 5975 120 - 1 CGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUUUGCUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUUGAGUCCUUCUUGG ((((((((((.........((((((..........))))))..((..((((((.((.((.......))....))..))))))..))....)))).....(((((...))))).)))))). ( -39.20) >DroEre_CAF1 7015 120 - 1 CAACAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCUGCCUCCAGAGCCUUCUUCUGCUCCUUCUGCUUCUGCUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUCGAGUCCUUCUUCG ......((((.(...(((.(((((((((......)).))))))))))((((((....((.............))..))))))...).))))((((...........)))).......... ( -36.72) >DroYak_CAF1 6148 120 - 1 CAACAAGUGGUCCCUUCUUGGAGGCACCAGCCUUGGCUGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUCGAGUCCUUCUUUG ......(.((.(...(((.(((((((((......)).))))))))))((((((.((.((.......))....))..))))))...).)).)((((...........)))).......... ( -35.10) >consensus CAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAGGCCAUGUCCUCCUCGUCUAGAUUCUUCGAGUCCUUCUUGG .(((((((((.(.....((((((((....((....)).)))))))).((((((.((.((.......))....))..))))))...).))))((((...........))))...))))).. (-31.00 = -31.88 + 0.88)

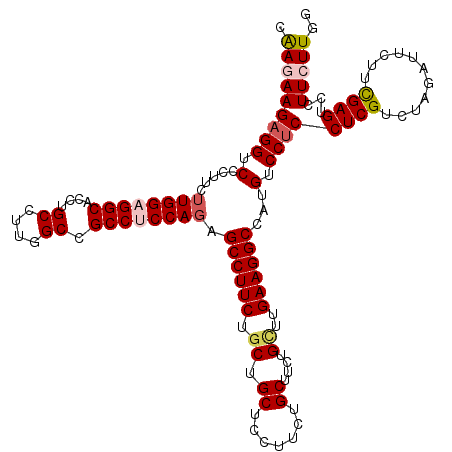
| Location | 13,293,161 – 13,293,281 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -32.28 |
| Energy contribution | -33.44 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293161 120 + 22407834 CUUCAAACAGAAGCAGAAGGAGCAGCAGAAGGCUAUGGAGGCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUGGCGGCGGUAUCAAAAAGUCCGGCAAAAAGUGAUCCAUUGC ((((.....)))).....(((...((.....((((((((((((.((......))))))))))((((((....))))))))))(.(((...........))).).....))..)))..... ( -37.30) >DroSec_CAF1 6030 120 + 1 CUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCGGCGGCGGAAUCAAGAAGUCCGGCAAAAAGUGAUCCAUAGC ((((..((....)).))))((((........))))((((.((.(((.........((((((.((((((....)))))).))))))(((.........)))))).....))..)))).... ( -41.50) >DroSim_CAF1 6015 120 + 1 CUUCAAGCAAAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCGGCGGCGGGAUCAAGAAGUCCGGCAAAAAGUGAUCCAUAGC ((((..((....)).))))((((........))))((((.((.(((........(((((((.((((((....)))))).)))))))((((......))))))).....))..)))).... ( -43.70) >DroEre_CAF1 7055 120 + 1 CUUCAAGCAGAAGCAGAAGGAGCAGAAGAAGGCUCUGGAGGCAGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUGUUGGCGGCGGGAUCAAAAAGUCGGGCAAAAAGUGAUCCCUAGC ((((..((....)).))))((((........))))((((((((.((......))))))))))...(((((.(.(((.(((.((((...........))).).))).))).).)))))... ( -40.80) >DroYak_CAF1 6188 120 + 1 CUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCAGCCAAGGCUGGUGCCUCCAAGAAGGGACCACUUGUUGGCGGCGGGAUCAAAAAGUCGGGCAAAAAGUGAUCCCUAGC ((((..((....)).))))((((........))))((((((((.(((....)))))))))))...(((((.(((((.(((.((((...........))).).))).))))).)))))... ( -45.70) >consensus CUUCAAGCAGAAGCAGAAGGAGCAGCAGAAGGCUCUGGAGGCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUGGCGGCGGGAUCAAAAAGUCCGGCAAAAAGUGAUCCAUAGC ((((..............(((((........)))))(((((((.((......)))))))))..))))(((.(.(((.(((.((((...........).))).))).))).).)))..... (-32.28 = -33.44 + 1.16)

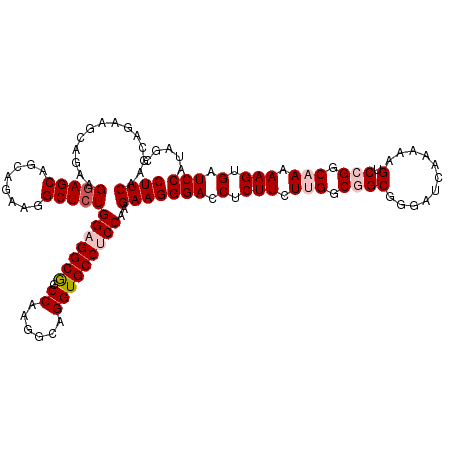
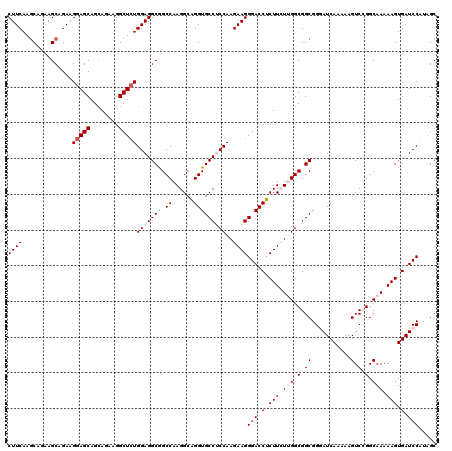
| Location | 13,293,161 – 13,293,281 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -40.31 |
| Consensus MFE | -32.56 |
| Energy contribution | -33.56 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293161 120 - 22407834 GCAAUGGAUCACUUUUUGCCGGACUUUUUGAUACCGCCGCCAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGCCUCCAUAGCCUUCUGCUGCUCCUUCUGCUUCUGUUUGAAG ((...(((((.(((((((.(((..............))).))))))).))))).....(((((((.((......))..)))))))..))((((.((.((.......))....))..)))) ( -37.04) >DroSec_CAF1 6030 120 - 1 GCUAUGGAUCACUUUUUGCCGGACUUCUUGAUUCCGCCGCCGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAG (((.((((.........((((((.........)))((((((((((((......)))))))))))).........)))....)))).)))((((.((.((.......))....))..)))) ( -41.32) >DroSim_CAF1 6015 120 - 1 GCUAUGGAUCACUUUUUGCCGGACUUCUUGAUCCCGCCGCCGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUUUGCUUGAAG (((.((((.........(((((((.....).))).((((((((((((......)))))))))))).........)))....)))).)))((((.((.((.......))....))..)))) ( -41.42) >DroEre_CAF1 7055 120 - 1 GCUAGGGAUCACUUUUUGCCCGACUUUUUGAUCCCGCCGCCAACAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCUGCCUCCAGAGCCUUCUUCUGCUCCUUCUGCUUCUGCUUGAAG ((..(((((((.................)))))))...))(((..((((((.......((((((((((......)).))))))))((((........)))).....))))))..)))... ( -39.23) >DroYak_CAF1 6188 120 - 1 GCUAGGGAUCACUUUUUGCCCGACUUUUUGAUCCCGCCGCCAACAAGUGGUCCCUUCUUGGAGGCACCAGCCUUGGCUGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAG ...(((((((((((.(((..((...............)).))).)))))))))))...((((((((((......)).))))))))((((........))))((((.((....))..)))) ( -42.56) >consensus GCUAUGGAUCACUUUUUGCCGGACUUUUUGAUCCCGCCGCCAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGCCUCCAGAGCCUUCUGCUGCUCCUUCUGCUUCUGCUUGAAG .....(((((.(((((((.(((..............))).))))))).))))).....(((((((....((....)).)))))))((((........))))((((.((....))..)))) (-32.56 = -33.56 + 1.00)

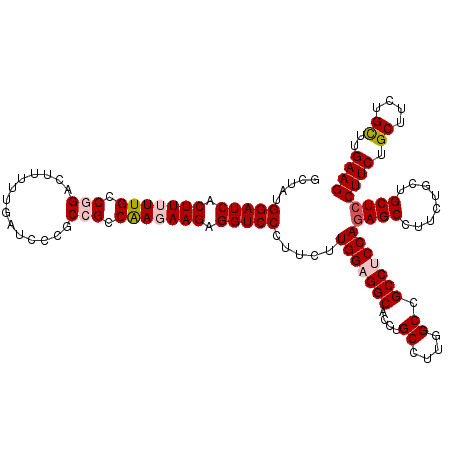
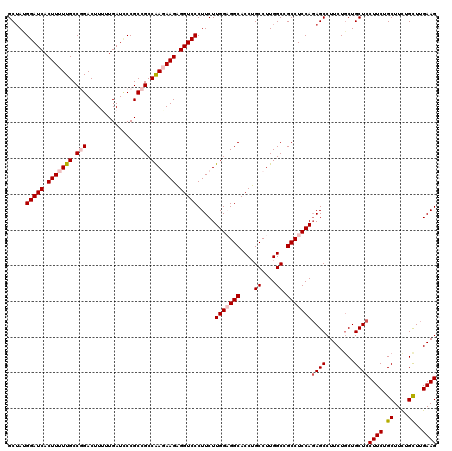
| Location | 13,293,201 – 13,293,313 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293201 112 + 22407834 GCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUGGCGGCGGUAUCAAAAAGUCCGGCAAAAAGUGAUCCAUUGCCACACCUUCAUCUGUAUAU--------CUUGGCAUAUUCA ...(((((((((((((((.(((((((((....))))))))).)))...............(((((...........)))))........))))))....--------))))))....... ( -39.50) >DroSec_CAF1 6070 112 + 1 GCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCGGCGGCGGAAUCAAGAAGUCCGGCAAAAAGUGAUCCAUAGCCACACUUUCAUAUGCAUUC--------CUUAGCAUAUUCA ((.(((......)))((((((.((((((....)))))).))))))(((((((.(((((..(((...............)))..)))))....)).))))--------)...))....... ( -35.96) >DroSim_CAF1 6055 112 + 1 GCGGCCAAGGCAGGUGCCGCCAAGAAGGGACCUCUUCUCGGCGGCGGGAUCAAGAAGUCCGGCAAAAAGUGAUCCAUAGCCACACUUUCAUAUGAAUUC--------CUUAGCAUAUUCA ...((.((((..(((((((((.((((((....)))))).)))))).((((((....(.....)......))))))...))).....(((....)))..)--------))).))....... ( -37.00) >DroEre_CAF1 7095 112 + 1 GCAGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUGUUGGCGGCGGGAUCAAAAAGUCGGGCAAAAAGUGAUCCCUAGCCGCGGCUUCAUCUGUUUUC--------CUUAACUGAAUUA (.((((..(((....(((.((((.((((....)))).)))).)))(((((((....(.....)......)))))))..)))..)))).).((.(((...--------...))).)).... ( -38.60) >DroYak_CAF1 6228 120 + 1 GCAGCCAAGGCUGGUGCCUCCAAGAAGGGACCACUUGUUGGCGGCGGGAUCAAAAAGUCGGGCAAAAAGUGAUCCCUAGCCACGCCUUCAUCUUCUUUUAAAUCGCACCCAUGUGGAUUA ......((((((((((((.((((.(((......))).)))).)))(((((((....(.....)......)))))))..)))).)))))............((((.(((....))))))). ( -40.30) >consensus GCGGCCAAGGCAGGUGCCUCCAAGAAGGGACCUCUUCUUGGCGGCGGGAUCAAAAAGUCCGGCAAAAAGUGAUCCAUAGCCACACCUUCAUCUGCAUUC________CUUAGCAUAUUCA ...(((((((.((((.((........)).))))..)))))))(((.((((((....(.....)......))))))...)))....................................... (-26.12 = -27.00 + 0.88)

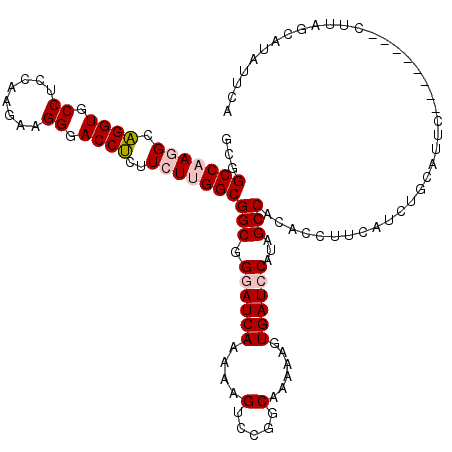
| Location | 13,293,201 – 13,293,313 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -28.29 |
| Energy contribution | -28.33 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13293201 112 - 22407834 UGAAUAUGCCAAG--------AUAUACAGAUGAAGGUGUGGCAAUGGAUCACUUUUUGCCGGACUUUUUGAUACCGCCGCCAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGC .......((((((--------.....(((..((((((.((((((.((....))..)))))).)))))).......(((.((((((((......)))))))).)))..))).))))))... ( -41.60) >DroSec_CAF1 6070 112 - 1 UGAAUAUGCUAAG--------GAAUGCAUAUGAAAGUGUGGCUAUGGAUCACUUUUUGCCGGACUUCUUGAUUCCGCCGCCGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGC .......((((((--------((((.((...(((.((.((((...((....))....)))).))))).)))))))((((((((((((......)))))))))))).......)))))... ( -44.30) >DroSim_CAF1 6055 112 - 1 UGAAUAUGCUAAG--------GAAUUCAUAUGAAAGUGUGGCUAUGGAUCACUUUUUGCCGGACUUCUUGAUCCCGCCGCCGAGAAGAGGUCCCUUCUUGGCGGCACCUGCCUUGGCCGC .......((((((--------(...(((...(((.((.((((...((....))....)))).))))).)))....((((((((((((......)))))))))))).....)))))))... ( -45.10) >DroEre_CAF1 7095 112 - 1 UAAUUCAGUUAAG--------GAAAACAGAUGAAGCCGCGGCUAGGGAUCACUUUUUGCCCGACUUUUUGAUCCCGCCGCCAACAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCUGC .....((((((((--------(....(((.....((((((((..(((((((.................))))))))))))...(((((((...)))))))..)))..)))))))))))). ( -40.73) >DroYak_CAF1 6228 120 - 1 UAAUCCACAUGGGUGCGAUUUAAAAGAAGAUGAAGGCGUGGCUAGGGAUCACUUUUUGCCCGACUUUUUGAUCCCGCCGCCAACAAGUGGUCCCUUCUUGGAGGCACCAGCCUUGGCUGC .(((((((....))).)))).......((.(.((((((((((..(((((((.................)))))))))))).......((((.((((....)))).))))))))).))).. ( -38.93) >consensus UGAAUAUGCUAAG________GAAUACAGAUGAAGGUGUGGCUAUGGAUCACUUUUUGCCGGACUUUUUGAUCCCGCCGCCAAGAAGAGGUCCCUUCUUGGAGGCACCUGCCUUGGCCGC .....................................(((((((.((((((.................)))))).(((.((((((((......)))))))).)))........))))))) (-28.29 = -28.33 + 0.04)

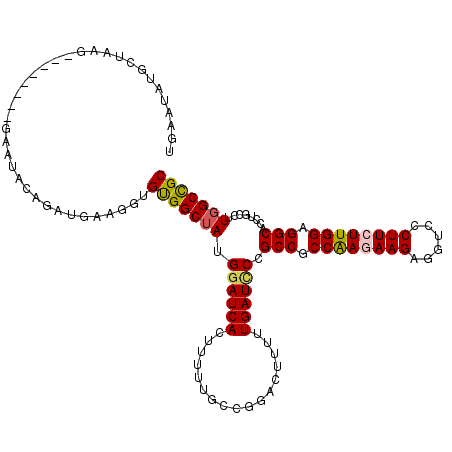
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:27 2006