
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,288,280 – 13,288,382 |
| Length | 102 |
| Max. P | 0.892316 |

| Location | 13,288,280 – 13,288,382 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
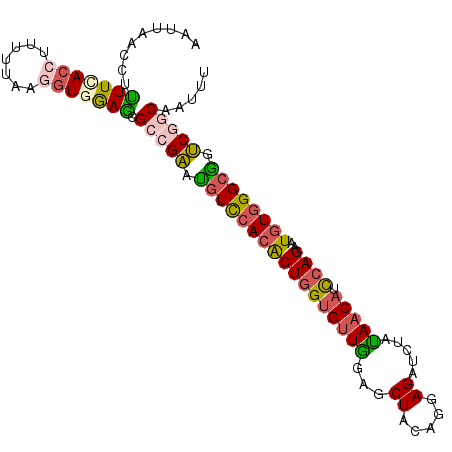
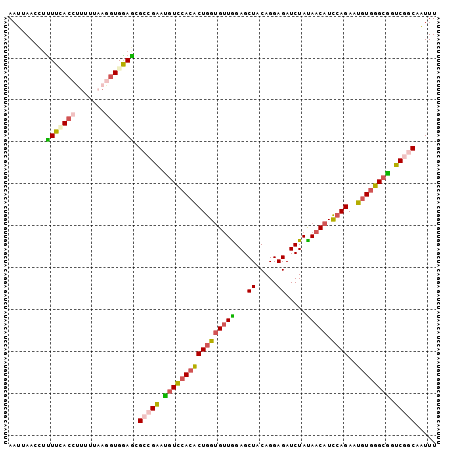
>2L_DroMel_CAF1 13288280 102 + 22407834 AAUAAACCUUUUCACCCUUUUAAGGUGGAGCGCCGAAUGUCCACACUGGUGUUGGAGCUACAGGAGAUCUAUAACAUCCAGAAUGUGGGCGGUCGGCAAUUU .........(((((((.......))))))).(((((.(((((((((((((((((((.((.....)).))))..))).))))..)))))))).)))))..... ( -37.80) >DroGri_CAF1 1609 100 + 1 AUCAAAUCGUUUAA--UAUUGCAGGUGAAGCGACGCAGAUUCACGCUGGCGCUGGAGCUGCAGGAGAUCUACAACAUUAAGAGUCUGGGCUCGCAACAGUUU .(((..(.((....--....)))..))).(((((.(((((((..((.(((......))))).((....))..........))))))).).))))........ ( -23.70) >DroSec_CAF1 1342 102 + 1 AAUUAAUCUUUUUACGUUUUUAAGGUGGAGCGCAGAAUGUCCACACUGGUGUUGGAGCUUCAGGAGAUCUAUAACAUCCAGAAUGUGGGCGGUCGGCAAUUU ...............(((((......)))))((.((.(((((((((((((((((.((..((....)).)).))))).))))..)))))))).)).))..... ( -28.50) >DroSim_CAF1 1340 102 + 1 AAUUAACCUCUUUACCUUUUUAAGGUGGAGCGCCGAAUGUCGACACUGGUGUUAGAGCUACAGGAGAUCUAUAACAUCCAGAAUGUGGGCGGUCGGCAAUUU .........((((((((.....)))))))).(((((.((((.((((((((((((((.((.....)).))))..))).))))..))).)))).)))))..... ( -35.10) >DroEre_CAF1 1337 102 + 1 AAUUAAGUUUUUCACCUUAAUUAGGUGGAGCGCCGAAUGUCCACACUGGUGUUGGAGCUACAGGAGAUCUAUAACAUCCAGAAUGUGGGCAGUCGGCAAUUU .........((((((((.....)))))))).(((((.(((((((((((((((((((.((.....)).))))..))).))))..)))))))).)))))..... ( -40.70) >DroYak_CAF1 1339 102 + 1 AAUGAACAUUUUGACCAUUCCAAGGUGGAACGUCGAAUGUCCACACUGGUGUUGGAGCUACAGGAGAUCUACAACAUCCAGAAUGUGGGCGGUCGUCAAUUU ......(((((((.......)))))))....(.(((.(((((((((((((((((...((.....)).....))))).))))..)))))))).))).)..... ( -31.80) >consensus AAUUAACCUUUUCACCUUUUUAAGGUGGAGCGCCGAAUGUCCACACUGGUGUUGGAGCUACAGGAGAUCUAUAACAUCCAGAAUGUGGGCGGUCGGCAAUUU .........(((((((.......))))))).(((((.(((((((((((((((((...((.....)).....))))).))))..)))))))).)))))..... (-26.25 = -26.78 + 0.54)

| Location | 13,288,280 – 13,288,382 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
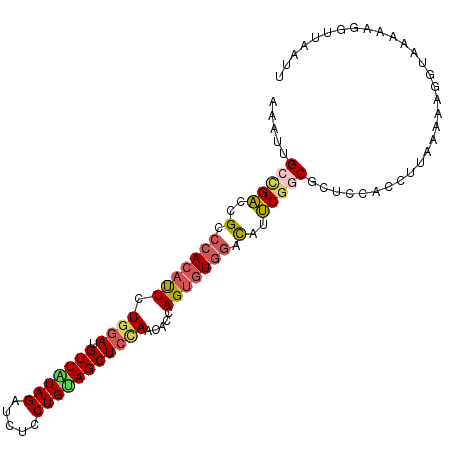
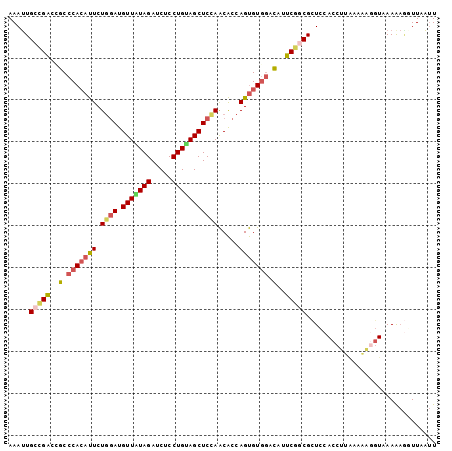
>2L_DroMel_CAF1 13288280 102 - 22407834 AAAUUGCCGACCGCCCACAUUCUGGAUGUUAUAGAUCUCCUGUAGCUCCAACACCAGUGUGGACAUUCGGCGCUCCACCUUAAAAGGGUGAAAAGGUUUAUU .....(((((..(.(((((((.((((.(((((((.....))))))))))).....))))))).)..)))))....(((((.....)))))............ ( -34.00) >DroGri_CAF1 1609 100 - 1 AAACUGUUGCGAGCCCAGACUCUUAAUGUUGUAGAUCUCCUGCAGCUCCAGCGCCAGCGUGAAUCUGCGUCGCUUCACCUGCAAUA--UUAAACGAUUUGAU ....(((((((((......))).....(((((((.....)))))))...((((...((((......))))))))......))))))--.............. ( -22.10) >DroSec_CAF1 1342 102 - 1 AAAUUGCCGACCGCCCACAUUCUGGAUGUUAUAGAUCUCCUGAAGCUCCAACACCAGUGUGGACAUUCUGCGCUCCACCUUAAAAACGUAAAAAGAUUAAUU .....((.((..(.(((((..((((.((((..((.((....))..))..))))))))))))).)..)).))............................... ( -18.10) >DroSim_CAF1 1340 102 - 1 AAAUUGCCGACCGCCCACAUUCUGGAUGUUAUAGAUCUCCUGUAGCUCUAACACCAGUGUCGACAUUCGGCGCUCCACCUUAAAAAGGUAAAGAGGUUAAUU ........((((..........((((.(((((((.....))))))))))).....(((((((.....)))))))..((((.....)))).....)))).... ( -24.80) >DroEre_CAF1 1337 102 - 1 AAAUUGCCGACUGCCCACAUUCUGGAUGUUAUAGAUCUCCUGUAGCUCCAACACCAGUGUGGACAUUCGGCGCUCCACCUAAUUAAGGUGAAAAACUUAAUU .....(((((.((.(((((((.((((.(((((((.....))))))))))).....))))))).)).)))))....(((((.....)))))............ ( -36.40) >DroYak_CAF1 1339 102 - 1 AAAUUGACGACCGCCCACAUUCUGGAUGUUGUAGAUCUCCUGUAGCUCCAACACCAGUGUGGACAUUCGACGUUCCACCUUGGAAUGGUCAAAAUGUUCAUU ...(((((((..(.(((((..((((.(((((.((..(....)...)).)))))))))))))).)..))..((((((.....))))))))))).......... ( -31.10) >consensus AAAUUGCCGACCGCCCACAUUCUGGAUGUUAUAGAUCUCCUGUAGCUCCAACACCAGUGUGGACAUUCGGCGCUCCACCUUAAAAAGGUAAAAAGGUUAAUU .....(((((..(.(((((((.((((.(((((((.....))))))))))).....))))))).)..)))))............................... (-19.72 = -20.33 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:21 2006