| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,285,549 – 13,285,654 |
| Length | 105 |
| Max. P | 0.549020 |

| Location | 13,285,549 – 13,285,654 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
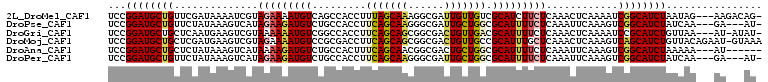
>2L_DroMel_CAF1 13285549 105 + 22407834 UCCGGAUGCUGUUCGAUAAAAUCGUAGAAAAUGUCAGCCACCUUUAGCAAAGGCGAUUGUUGUCGCAUCUUCUCAAACUCAAAAUCGGCAUCUAAUAG---AAGACAG- ...((((((((..((((...)))).((((.(((((((((.((((.....)))).)...))))..)))).))))............)))))))).....---.......- ( -20.90) >DroPse_CAF1 22218 102 + 1 UCCGGAUGCUGUUCUAUAAAGUCAUAGAAGAUGUCUGCCACCUUCAGCAAGGGCGAUUGCUGGCGCAUUUUCUCAAAUUCAAAGUCGGCAUCUAUCAA---GA---AU- ...((((((((..((..........(((((((((..((((((((....))))((....))))))))))))))).........)).)))))))).....---..---..- ( -27.61) >DroGri_CAF1 21534 104 + 1 UCCGGAUGCUGCUCAAUGAAGUCGUAAAAAAUGUCGGCCACCUUCAGCAGCGGCGACUGUUGACGCAUUUUCUCAAACUCAAAAUCCGCAUCUGUUAA---AU-AUAU- ..(((((((.(((((((..((((((......((..((...))..))......))))))))))).))(((((.........)))))..)))))))....---..-....- ( -23.10) >DroMoj_CAF1 25774 108 + 1 UCCGGAUGCUGCUCGAUGAAGUCGUAGAAAAUGUCCGCGACCUUCAGCAGCGGCGACUGUUGCCGCAUUUGCUCAAACUCAAAGUCAGCAUCUGUUACAGAAU-GUAAA ..(((((((((((...((((((((..((.....))..))).)))))((((((((((...)))))))...)))..........)).)))))))))((((.....-)))). ( -38.20) >DroAna_CAF1 20776 100 + 1 UCCGGAUGCUGCUCUAUAAAGUCAUAAAAGAUGUCUGCCACUUUCAGCAACGGCGACUGCUGGCGCAUUUUCUCAAAUUCAAAGUCGGCAUCUAAAAA---AU------ ...((((((((..((............(((((((..(((((..((.((....))))..).)))))))))))...........)).)))))))).....---..------ ( -22.30) >DroPer_CAF1 22953 102 + 1 UCCGGAUGCUGUUCUAUAAAGUCAUAGAAGAUGUCUGCCACCUUCAGCAAGGGCGAUUGCUGGCGCAUUUUCUCAAAUUCAAAGUCGGCAUCUAUCAA---GA---AU- ...((((((((..((..........(((((((((..((((((((....))))((....))))))))))))))).........)).)))))))).....---..---..- ( -27.61) >consensus UCCGGAUGCUGCUCUAUAAAGUCAUAGAAAAUGUCUGCCACCUUCAGCAACGGCGACUGCUGGCGCAUUUUCUCAAACUCAAAGUCGGCAUCUAUUAA___AA___AU_ ...((((((((..............(((((((((.........(((((((......))))))).)))))))))............))))))))................ (-19.14 = -19.39 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:17 2006