
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,256,092 – 13,256,219 |
| Length | 127 |
| Max. P | 0.969470 |

| Location | 13,256,092 – 13,256,201 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
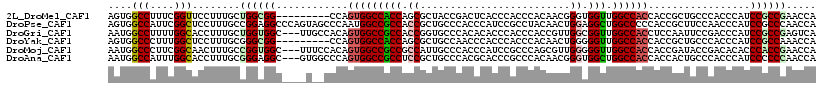
>2L_DroMel_CAF1 13256092 109 + 22407834 AGUGGCCUUUCGGUUCCUUUGCUGGCGG---------CCAGUGGCCACCAGCGCUACCGACUCACCCACCCACAACGGGUGGUUGGCCACCACCGCUGCCCACCCAUCCGCCGAACCA .((((((..(((((.....((((((.((---------((...)))).))))))..))))).....((((((.....))))))..))))))............................ ( -46.00) >DroPse_CAF1 40829 118 + 1 AGUGGCCAUUCGGCUCCUUUGCCGGAGGCCCAGUAGCCCAAUGGCCGCCACCGCUGCCCACCCAUCCGCCUACAACUGGAGGCUGGCCCCCACCGCUUCCAACCCAUCCGCCCAACCA .(((((((((.((((.((..(((...)))..)).)))).)))))))))............................(((((((.((......)))))))))................. ( -38.50) >DroGri_CAF1 31481 115 + 1 AAUGGCCUUUUGGCACCUUUGCUGGUGGC---UUGCCACAGUGGCCGCCACCGGUGCCCACACACCCACCCACCGUUGGCGGUUGGCCACCUCCAAUUCCGACCCAUCCGCCGAGUCA ..((((.....((((((......((((((---..(((.....))).)))))))))))).................(((((((.(((.................))).))))))))))) ( -41.63) >DroYak_CAF1 31139 109 + 1 AGUGGCCCUUUGGCUCCUUUGCGGGCGG---------CCAGUGGCCACCAGCGCUGCCAACCCACCCACCCACAACUGGGGGUUGGCCACCACCGCUGCCCACCCAUCCGCCAAACCA .((((((..(((((((((....))).))---------)))).))))))(((((..((((((((..(((........)))))))))))......))))).................... ( -44.40) >DroMoj_CAF1 38365 115 + 1 AAUGGCCCUUCGGCAACUUUGCCGGUGGC---UUUCCACAGUGGCCGCCGCCAUUGCCCACCCAUCCGCCCAGCGUUGGGGGUUGGCCACCACCGAUACCGACACACCCACCGAACCA ..(((...(((((((....))))((((((---(..((.(((((((....)))))))....((((..(((...))).))))))..))))))).....................)))))) ( -43.40) >DroAna_CAF1 38603 115 + 1 AAUGGCCAUUUGGCACCUUUGCGGGAGGC---GUGGCCCAGUGGCCGCCUCCGCUGCCCACGCACCCGCCCACAACGGGUGGCUGGCCACCACCACUGCCCACCCAUCCCCCCAACCA ....(((....)))......(((((..((---((((..((((((......)))))).)))))).))))).......((((((((((......)))..).))))))............. ( -47.70) >consensus AAUGGCCAUUCGGCACCUUUGCCGGAGGC____U__CCCAGUGGCCGCCACCGCUGCCCACCCACCCACCCACAACUGGGGGUUGGCCACCACCGCUGCCCACCCAUCCGCCGAACCA ....(((....)))........((((((............(((((((((.((.........................)).))).)))))).................))))))..... (-22.62 = -22.96 + 0.34)
| Location | 13,256,092 – 13,256,201 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -54.77 |
| Consensus MFE | -30.27 |
| Energy contribution | -31.05 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
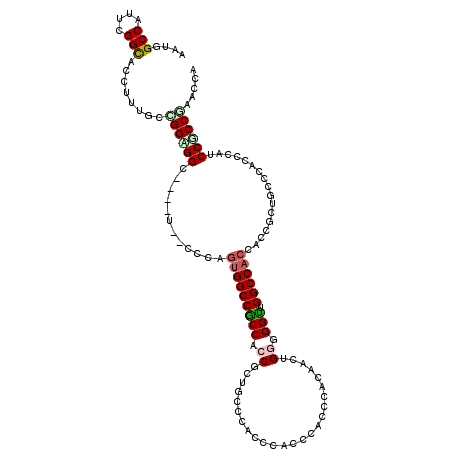
>2L_DroMel_CAF1 13256092 109 - 22407834 UGGUUCGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCACCCGUUGUGGGUGGGUGAGUCGGUAGCGCUGGUGGCCACUGG---------CCGCCAGCAAAGGAACCGAAAGGCCACU ..(((.(((((..((((.(.(((((.((.((((((.((((((.....)))))).)).)))).)).))))).).))).)...---------))))))))...((..((....))))... ( -55.00) >DroPse_CAF1 40829 118 - 1 UGGUUGGGCGGAUGGGUUGGAAGCGGUGGGGGCCAGCCUCCAGUUGUAGGCGGAUGGGUGGGCAGCGGUGGCGGCCAUUGGGCUACUGGGCCUCCGGCAAAGGAGCCGAAUGGCCACU ((((((..(((....((.....))(.((((((((.(((.((.(((((..((......))..))))))).)))((((....))))....)))))))).).......)))..)))))).. ( -49.80) >DroGri_CAF1 31481 115 - 1 UGACUCGGCGGAUGGGUCGGAAUUGGAGGUGGCCAACCGCCAACGGUGGGUGGGUGUGUGGGCACCGGUGGCGGCCACUGUGGCAA---GCCACCAGCAAAGGUGCCAAAAGGCCAUU .((((((.....)))))).........(((((((..(((((.......))))).......((((((((((((.(((.....)))..---))))))......))))))....))))))) ( -52.60) >DroYak_CAF1 31139 109 - 1 UGGUUUGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCCCCAGUUGUGGGUGGGUGGGUUGGCAGCGCUGGUGGCCACUGG---------CCGCCCGCAAAGGAGCCAAAGGGCCACU ..((..(((((.(.(((((.((.(((((.((.((((((((((.(....).)))..))))))))).))))).)).))))).)---------))))).))..((..(((....)))..)) ( -55.90) >DroMoj_CAF1 38365 115 - 1 UGGUUCGGUGGGUGUGUCGGUAUCGGUGGUGGCCAACCCCCAACGCUGGGCGGAUGGGUGGGCAAUGGCGGCGGCCACUGUGGAAA---GCCACCGGCAAAGUUGCCGAAGGGCCAUU ((((((((..(.(.(((((((..(((((((.(((..((((((.(((...)))..)))).))((....))))).))))))).((...---.))))))))).).)..))))...)))).. ( -57.00) >DroAna_CAF1 38603 115 - 1 UGGUUGGGGGGAUGGGUGGGCAGUGGUGGUGGCCAGCCACCCGUUGUGGGCGGGUGCGUGGGCAGCGGAGGCGGCCACUGGGCCAC---GCCUCCCGCAAAGGUGCCAAAUGGCCAUU ((((((..((.(((((.((((.((((((((((((.(((..(((((((..(((....)))..))))))).))))))))))..)))))---)))))))......)).))...)))))).. ( -58.30) >consensus UGGUUCGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCACCAGUUGUGGGCGGGUGGGUGGGCAGCGGUGGCGGCCACUGGG__A____GCCACCAGCAAAGGAGCCAAAAGGCCACU (((((((((......((((((..(((((((.(((..((.((.......)).)).((......)).....))).)))))))((........))))))))......)))...)))))).. (-30.27 = -31.05 + 0.78)

| Location | 13,256,123 – 13,256,219 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -28.56 |
| Energy contribution | -28.20 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
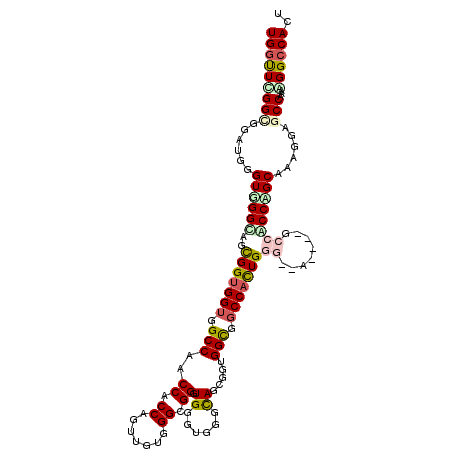
>2L_DroMel_CAF1 13256123 96 - 22407834 GGGUGCGUGGGAUAAUGGUGGUUCGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCACCCGUUGUGGGUGGGUGAGUCGGUAGCGCUGGUGGCCAC ..((.(((.((((.......)))).))).)).(((.(.(((((.((.((((((.((((((.....)))))).)).)))).)).))))).).))).. ( -44.70) >DroVir_CAF1 35705 96 - 1 GGAUGCGUUGGAUACUGAUGACUCGGAGGAUGAGUCGGUAUCGGAGGUGGCCAACCGCCAACGGUGGGCGGAUGCGUCGGUAUUGGUGGCGGCCAC ....((.((.((((((...((((((.....)))))))))))).)).))((((..(((((((...(.((((....)))).)..))))))).)))).. ( -39.80) >DroGri_CAF1 31518 96 - 1 GGAUGCGUGGGAUACUGAUGACUCGGCGGAUGGGUCGGAAUUGGAGGUGGCCAACCGCCAACGGUGGGUGGGUGUGUGGGCACCGGUGGCGGCCAC ......((((..((((..(((((((.....)))))))........((((.(((..((((.((.....)).))))..))).))))))))....)))) ( -34.00) >DroEre_CAF1 30885 96 - 1 GGAUGCGUGGGAUAAUGGUGGUUCGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCGCCCGUGGUGGGUGGGUGCGUCGGUAGCGCUGGUGGCCAC ..((.(((.((((.......)))).))).)).(((.(.(((((.((.((((((.((((((.....)))))).)).)))).)).))))).).))).. ( -44.00) >DroYak_CAF1 31170 96 - 1 GGAUGCGUAGGAUAAUGGUGGUUUGGCGGAUGGGUGGGCAGCGGUGGUGGCCAACCCCCAGUUGUGGGUGGGUGGGUUGGCAGCGCUGGUGGCCAC ..((.(((..(((.......)))..))).))..((((.((.(((((.((.((((((((((.(....).)))..))))))))).))))).)).)))) ( -40.30) >DroMoj_CAF1 38402 96 - 1 GGAUGCGUUGGAUACUGAUGGUUCGGUGGGUGUGUCGGUAUCGGUGGUGGCCAACCCCCAACGCUGGGCGGAUGGGUGGGCAAUGGCGGCGGCCAC ..........((((((((((..((....))..))))))))))(((.((.((((.((((((.(((...)))..)))).))....)))).)).))).. ( -39.20) >consensus GGAUGCGUGGGAUAAUGAUGGUUCGGCGGAUGGGUCGGCAGCGGUGGUGGCCAACCGCCAACGGUGGGUGGGUGCGUCGGCAGCGCUGGCGGCCAC ..((.(((.((((.......)))).))).)).(((.(.(((((.((.((((((.(((((.......))))).)).)))).)).))))).).))).. (-28.56 = -28.20 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:23:13 2006