| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,224,732 – 13,224,850 |
| Length | 118 |
| Max. P | 0.866904 |

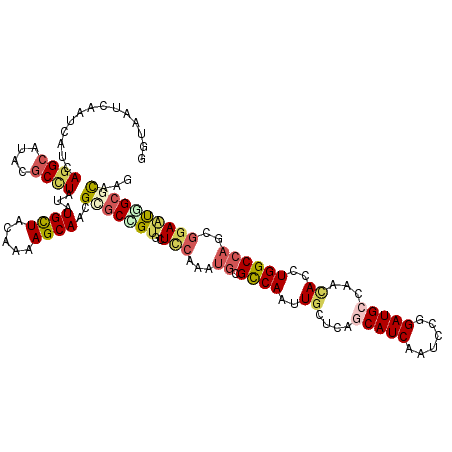
| Location | 13,224,732 – 13,224,850 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -43.46 |
| Consensus MFE | -21.94 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13224732 118 + 22407834 CUGGCGCCAUUCCAGUGGCCAGGUGUUGGCAUCCGGAUUGAUGUUGAGCAGUUGACGCAUCUGGAGCACGGCACGUUGCUUUUGUAGCAUGUAAGCGUAUGCCUGGUGAUUGAAGACC ............((((.(((((((((((((.(((((((.(.(((..(....)..))))))))))))).))))((((((((.....)))).....))))..))))))).))))...... ( -41.00) >DroVir_CAF1 1039 118 + 1 AUUGCGCAACUCCGCUGGCCAGGUGUUGGCAUCCGGAUUGAUGCUGAGCAUGUGGCGUAUUUGGAGUACCGCGCGUUGCUUUUGCAGCACAUAGGCGAACGCCUGAUGAUUCAUUAUC ...((((.((((((...((((.(((((((((((......)))))).))))).)))).....))))))...))))(((((....))))).(((((((....)))).))).......... ( -48.90) >DroGri_CAF1 1404 118 + 1 GUUGCGCAGUUCCGCUGGCCAGGUGUUGGCAUCCGGAUUGAUGCUCAGCAACUGGCGCAUUUGAAGCACGGCGCGCUGCUUCUGUAACACAUAGGCGAAUGCCUGAUGAUUGAUUACU ((((((((((..((((((((((.((((((((((......))))).))))).)))))((.......)).))))).)))))....))))).(((((((....)))).))).......... ( -48.90) >DroWil_CAF1 1285 115 + 1 UUGACGCCAUUCCAAUGGCCAAGUAUUUGCAUCCGGAUUGAUGCUGAGCAAUUGGCGCAUUUGAAGGACAGCUCUUUGCUUUUGUAGCAUCAAUGAGUAGGCUUGAUGAUUAAUA--- .....(((((....)))))(((((...(((.((...(((((((((((((((..(((..............)))..))))))....)))))))))))))).)))))..........--- ( -32.04) >DroMoj_CAF1 1124 118 + 1 AUUGCGCAAAUGUGCUGGCCAGGUAUUUGCAUCCGGGUUGAUGCUGAGCAAUUGGCGCAUCUGGAGCACGGUGCGUUGCUUUUGCAGCAUGUAGGCGAAUGCCUGAUGAUUGAUGCUU ...((((....))))....(((((((((((.(((((((.(.(((..(....)..)))))))))))((.....))(((((....)))))......)))))))))))............. ( -45.40) >DroAna_CAF1 2054 118 + 1 CUGGCGCCAUUCCACGGGCCAGGUGUUGGCAUCAGGAUUGAUGGCGAGGAGUUGGCGCAUCUGGAGCACAGCCCGUUGCUUCUGCAGCAUAUAGGCGUAGGCCUGAUGGCUAAAAACC (((((.((.......)))))))...(((((((((((....(((.(..((.((((((.(.....).)).))))))(((((....))))).....).)))...)))))).)))))..... ( -44.50) >consensus AUGGCGCAAUUCCACUGGCCAGGUGUUGGCAUCCGGAUUGAUGCUGAGCAAUUGGCGCAUCUGGAGCACGGCGCGUUGCUUUUGCAGCAUAUAGGCGAAUGCCUGAUGAUUGAUAACC .....((..........(((((...((((((((......))))))))....)))))(((...((((((........))))))))).))...(((((....)))))............. (-21.94 = -22.58 + 0.64)

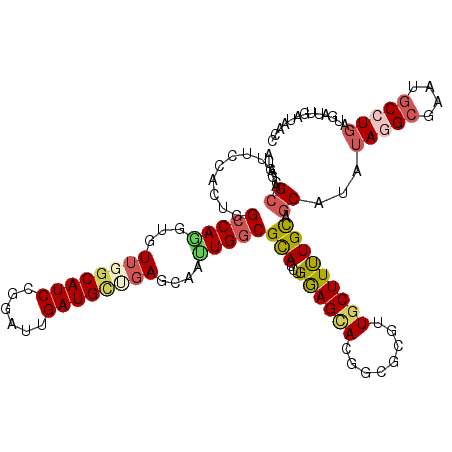
| Location | 13,224,732 – 13,224,850 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13224732 118 - 22407834 GGUCUUCAAUCACCAGGCAUACGCUUACAUGCUACAAAAGCAACGUGCCGUGCUCCAGAUGCGUCAACUGCUCAACAUCAAUCCGGAUGCCAACACCUGGCCACUGGAAUGGCGCCAG (((........))).(((...........((((.....))))....(((((..(((((.((.((((..((.....((((......))))....))..))))))))))))))))))).. ( -32.70) >DroVir_CAF1 1039 118 - 1 GAUAAUGAAUCAUCAGGCGUUCGCCUAUGUGCUGCAAAAGCAACGCGCGGUACUCCAAAUACGCCACAUGCUCAGCAUCAAUCCGGAUGCCAACACCUGGCCAGCGGAGUUGCGCAAU ..........(((.((((....)))))))((((.....))))..(((((..(((((......((((..((....(((((......)))))...))..))))....))))))))))... ( -36.80) >DroGri_CAF1 1404 118 - 1 AGUAAUCAAUCAUCAGGCAUUCGCCUAUGUGUUACAGAAGCAGCGCGCCGUGCUUCAAAUGCGCCAGUUGCUGAGCAUCAAUCCGGAUGCCAACACCUGGCCAGCGGAACUGCGCAAC .(((((....(((.((((....))))))).))))).......((((.((((((.......))(((((.((.((.(((((......))))))).)).)))))..))))....))))... ( -42.50) >DroWil_CAF1 1285 115 - 1 ---UAUUAAUCAUCAAGCCUACUCAUUGAUGCUACAAAAGCAAAGAGCUGUCCUUCAAAUGCGCCAAUUGCUCAGCAUCAAUCCGGAUGCAAAUACUUGGCCAUUGGAAUGGCGUCAA ---......................(((((((((....(((.....)))....(((.((((.(((((.......(((((......)))))......))))))))).)))))))))))) ( -28.82) >DroMoj_CAF1 1124 118 - 1 AAGCAUCAAUCAUCAGGCAUUCGCCUACAUGCUGCAAAAGCAACGCACCGUGCUCCAGAUGCGCCAAUUGCUCAGCAUCAACCCGGAUGCAAAUACCUGGCCAGCACAUUUGCGCAAU ..((..........((((....))))...((((.....))))..(((..(((((((((.((.((.....)).))(((((......)))))......))))..)))))...)))))... ( -35.00) >DroAna_CAF1 2054 118 - 1 GGUUUUUAGCCAUCAGGCCUACGCCUAUAUGCUGCAGAAGCAACGGGCUGUGCUCCAGAUGCGCCAACUCCUCGCCAUCAAUCCUGAUGCCAACACCUGGCCCGUGGAAUGGCGCCAG ((....(((((...((((....))))...((((.....))))...)))))....))....((((((..(((.((..((((....))))((((.....)))).)).))).))))))... ( -40.60) >consensus GGUAAUCAAUCAUCAGGCAUACGCCUAUAUGCUACAAAAGCAACGCGCCGUGCUCCAAAUGCGCCAAUUGCUCAGCAUCAAUCCGGAUGCCAACACCUGGCCAGCGGAAUGGCGCAAG ..............((((....))))...((((.....))))..(((((((..(((...((.((((..((....(((((......)))))...))..))))))..))))))))))... (-20.81 = -21.90 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:58 2006