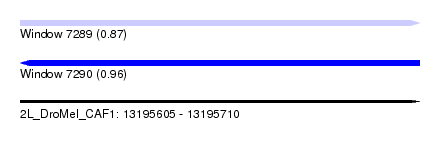
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,195,605 – 13,195,710 |
| Length | 105 |
| Max. P | 0.956165 |

| Location | 13,195,605 – 13,195,710 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -17.92 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
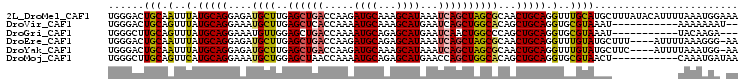
>2L_DroMel_CAF1 13195605 105 + 22407834 UGGGACUGCAAUUUAUGCAGGAGAUGCUUGAGCUGACCAAGAUGCAAAGCAUAAAUCAGCUAGCGCAACUGCAGGUUUGCAUGCUUUAUACAUUUUAAAUGGAAA .(((..(((((....(((((..(.((((..((((((.....((((...))))...)))))))))))..)))))...)))))..)))................... ( -28.40) >DroVir_CAF1 2162 92 + 1 UGGGACUGCAGUUUAUGCAGGAAAUGCUUGAGCUCACCAAAAUGCAAAGCAUGAAUCAGCUGGCACAGCUGCAGGUGCGUAAAU-----------AAAAAAAU-- ..........(((((((((....((((((..((..........)).))))))....((((((...))))))....)))))))))-----------........-- ( -23.00) >DroGri_CAF1 2267 91 + 1 UGGGCUUGCAGUUUAUGCAGGAAAUGUUGGAGCUGACCAAAAUGCAGAGCAUGAAUCAACUGGCCCAGCUGCAGGUGCGUAAAU-----------UACAAGA--- ....((((.((((((((((......(((((.(((.......((((...)))).........))))))))......)))))))))-----------).)))).--- ( -26.09) >DroEre_CAF1 5875 100 + 1 UGGGACUGCAAUUUAUGCAGGAGAUGCUUGAGCUGACCAAGAUGCAGAGCAUAAAUCAGCUAGCGCAACUGCAGGUUUGUAUGCUUU----AUUUUAAAGGG-AA .(((..(((((....(((((..(.((((..((((((.....((((...))))...)))))))))))..)))))...)))))..))).----...........-.. ( -26.40) >DroYak_CAF1 2937 100 + 1 UGGGACUGCAAUUUAUGCAGGAGAUGCUUGAGCUGACCAAGAUGCAAAGCAUAAAUCAGCUAGCGCAACUGCAGGUUUGUAUGCUUC----AUUUUAAAUGG-AA (((..((((((....(((((..(.((((..((((((.....((((...))))...)))))))))))..)))))...))))).)..))----)..........-.. ( -27.00) >DroMoj_CAF1 2166 94 + 1 UGGGCUUGCAGUUCAUGCAGGAAAUGCUGGAGCUAACCAAAAUGCAGAGCAUGAACCAGCUGGCACAGCUGCAGGUGCGUAACU-----------CAAAUGAUAA .(.((((((.((((((((......((((((......)))....)))..)))))))).(((((...))))))))))).)......-----------.......... ( -28.80) >consensus UGGGACUGCAAUUUAUGCAGGAAAUGCUUGAGCUGACCAAAAUGCAAAGCAUAAAUCAGCUAGCGCAACUGCAGGUGCGUAAGC___________UAAAUGA_AA ......(((.(..(.(((((....((((..((((((.....((((...))))...))))))))))...))))).)..))))........................ (-17.92 = -17.42 + -0.50)

| Location | 13,195,605 – 13,195,710 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -18.17 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13195605 105 - 22407834 UUUCCAUUUAAAAUGUAUAAAGCAUGCAAACCUGCAGUUGCGCUAGCUGAUUUAUGCUUUGCAUCUUGGUCAGCUCAAGCAUCUCCUGCAUAAAUUGCAGUCCCA .....................(((((((....))))..)))(((((((((((.((((...))))...))))))))..))).....(((((.....)))))..... ( -27.40) >DroVir_CAF1 2162 92 - 1 --AUUUUUUU-----------AUUUACGCACCUGCAGCUGUGCCAGCUGAUUCAUGCUUUGCAUUUUGGUGAGCUCAAGCAUUUCCUGCAUAAACUGCAGUCCCA --........-----------.......((((..((((((...))))))....((((...))))...)))).((....)).....(((((.....)))))..... ( -20.40) >DroGri_CAF1 2267 91 - 1 ---UCUUGUA-----------AUUUACGCACCUGCAGCUGGGCCAGUUGAUUCAUGCUCUGCAUUUUGGUCAGCUCCAACAUUUCCUGCAUAAACUGCAAGCCCA ---.......-----------......((....))....((((.((((((((.((((...))))...))))))))...........((((.....)))).)))). ( -20.70) >DroEre_CAF1 5875 100 - 1 UU-CCCUUUAAAAU----AAAGCAUACAAACCUGCAGUUGCGCUAGCUGAUUUAUGCUCUGCAUCUUGGUCAGCUCAAGCAUCUCCUGCAUAAAUUGCAGUCCCA ..-...........----...(((........)))((.(((...((((((((.((((...))))...))))))))...))).)).(((((.....)))))..... ( -24.60) >DroYak_CAF1 2937 100 - 1 UU-CCAUUUAAAAU----GAAGCAUACAAACCUGCAGUUGCGCUAGCUGAUUUAUGCUUUGCAUCUUGGUCAGCUCAAGCAUCUCCUGCAUAAAUUGCAGUCCCA ..-...........----...(((........)))((.(((...((((((((.((((...))))...))))))))...))).)).(((((.....)))))..... ( -24.60) >DroMoj_CAF1 2166 94 - 1 UUAUCAUUUG-----------AGUUACGCACCUGCAGCUGUGCCAGCUGGUUCAUGCUCUGCAUUUUGGUUAGCUCCAGCAUUUCCUGCAUGAACUGCAAGCCCA ..........-----------......(((....((((((...))))))((((((((........((((......))))........)))))))))))....... ( -25.09) >consensus UU_CCAUUUA___________ACAUACAAACCUGCAGCUGCGCCAGCUGAUUCAUGCUCUGCAUCUUGGUCAGCUCAAGCAUCUCCUGCAUAAACUGCAGUCCCA ................................(((((.(((...((((((((.((((...))))...))))))))...)))....)))))............... (-18.17 = -17.95 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:41 2006