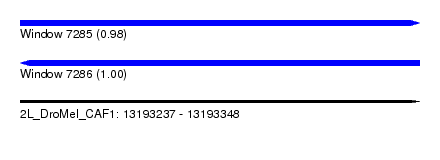
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,193,237 – 13,193,348 |
| Length | 111 |
| Max. P | 0.998895 |

| Location | 13,193,237 – 13,193,348 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -20.66 |
| Energy contribution | -22.26 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13193237 111 + 22407834 AAUUUGUUUAAAUGUAACCCAUAUAUGCGUCAGAAACGACGAGGUCAAAUUAGCAUAACUCGUUUUUAAUCAUUUAUGGAUAUACAUUUAAAUGUGAUGCUUAUAAAAACG--------- .....(((((((((((..(((((.(((.((.((((((((....((.......)).....)))))))).))))).)))))...)))))))))))..................--------- ( -22.30) >DroSec_CAF1 173 109 + 1 AAUUUGUUUAAAUGUAACCCAUAUAUGCAUCAGAAACGAGGAGGUCAAAUUAGCAUAACUCGUUUUUAAUCAUUUAUGGAUAUACAUUUAAAUGACAUGUUU--UAAAGCG--------- ...(..((((((((((..(((((.(((.((.(((((((((...((.......))....))))))))).))))).)))))...))))))))))..).......--.......--------- ( -27.90) >DroSim_CAF1 173 111 + 1 UAUUUGUUUAAAUGUAACCCAUAUAUGCAUCAGAAACGAGGAGGUCAAAUUAGCAUAACUCGUUUUUAAUCAUUUAUGGAUAUACAUUUAAAUGGCAUGCUUAUAAAAACG--------- ...(..((((((((((..(((((.(((.((.(((((((((...((.......))....))))))))).))))).)))))...))))))))))..)................--------- ( -27.90) >DroEre_CAF1 3478 120 + 1 AAUUUAUUUAAAUGUAACCCAUAUAAGCAUCAGAAACGAGGAGGUCAAAUUAGCAUAACUCGUUGGUAAUCAUUUAUGGUUUUUAAUUUAAAUGUGGUGCUUAUAAAUACCACUAUAAAA .....((((((((..((.(((((...........((((((...((.......))....))))))..........))))).))...))))))))((((((........))))))....... ( -22.60) >DroYak_CAF1 565 103 + 1 AACUUAUUUAAAUCUAACCCAUAUAUGCAUCAGAAACGAGGAGGUCUAAUUAGCAUAACUCGUUGGUGAUCAUUUAUGGUUAUAAAUUUA-AUGUGGUGCUUGU---------------- .(((...((((((.((..(((((.(((.((((..((((((...((.......))....))))))..))))))).)))))...)).)))))-)...)))......---------------- ( -23.50) >consensus AAUUUGUUUAAAUGUAACCCAUAUAUGCAUCAGAAACGAGGAGGUCAAAUUAGCAUAACUCGUUUUUAAUCAUUUAUGGAUAUACAUUUAAAUGUGAUGCUUAUAAAAACG_________ ....((((((((((((..(((((.(((.((.(((((((((...((.......))....))))))))).))))).)))))...)))))))))))).......................... (-20.66 = -22.26 + 1.60)

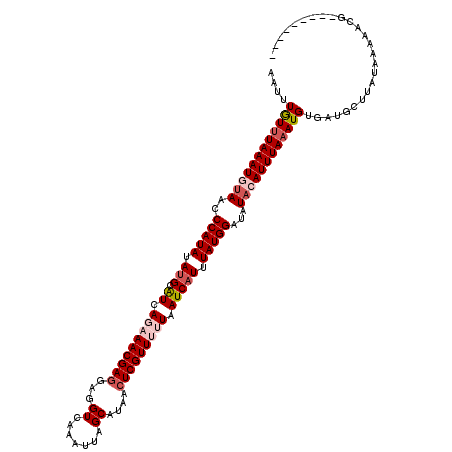
| Location | 13,193,237 – 13,193,348 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.60 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13193237 111 - 22407834 ---------CGUUUUUAUAAGCAUCACAUUUAAAUGUAUAUCCAUAAAUGAUUAAAAACGAGUUAUGCUAAUUUGACCUCGUCGUUUCUGACGCAUAUAUGGGUUACAUUUAAACAAAUU ---------.((((....))))......((((((((((.(((((((.(((.(((.((((((((((........))))....)))))).)))..))).)))))))))))))))))...... ( -25.30) >DroSec_CAF1 173 109 - 1 ---------CGCUUUA--AAACAUGUCAUUUAAAUGUAUAUCCAUAAAUGAUUAAAAACGAGUUAUGCUAAUUUGACCUCCUCGUUUCUGAUGCAUAUAUGGGUUACAUUUAAACAAAUU ---------.......--..........((((((((((.(((((((.(((((((.(((((((.....(......).....))))))).)))).))).)))))))))))))))))...... ( -27.60) >DroSim_CAF1 173 111 - 1 ---------CGUUUUUAUAAGCAUGCCAUUUAAAUGUAUAUCCAUAAAUGAUUAAAAACGAGUUAUGCUAAUUUGACCUCCUCGUUUCUGAUGCAUAUAUGGGUUACAUUUAAACAAAUA ---------.((((....))))......((((((((((.(((((((.(((((((.(((((((.....(......).....))))))).)))).))).)))))))))))))))))...... ( -28.80) >DroEre_CAF1 3478 120 - 1 UUUUAUAGUGGUAUUUAUAAGCACCACAUUUAAAUUAAAAACCAUAAAUGAUUACCAACGAGUUAUGCUAAUUUGACCUCCUCGUUUCUGAUGCUUAUAUGGGUUACAUUUAAAUAAAUU .......(((((..........)))))(((((((((((...(((((...(((((..((((((.....(......).....))))))..)))).)...))))).))).))))))))..... ( -21.70) >DroYak_CAF1 565 103 - 1 ----------------ACAAGCACCACAU-UAAAUUUAUAACCAUAAAUGAUCACCAACGAGUUAUGCUAAUUAGACCUCCUCGUUUCUGAUGCAUAUAUGGGUUAGAUUUAAAUAAGUU ----------------............(-((((((((...(((((.(((((((..((((((.....((....)).....))))))..)))).))).)))))..)))))))))....... ( -24.30) >consensus _________CGUUUUUAUAAGCAUCACAUUUAAAUGUAUAUCCAUAAAUGAUUAAAAACGAGUUAUGCUAAUUUGACCUCCUCGUUUCUGAUGCAUAUAUGGGUUACAUUUAAACAAAUU ............................((((((((((.(((((((.(((((((.(((((((.....(......).....))))))).)))).))).)))))))))))))))))...... (-21.00 = -22.60 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:38 2006