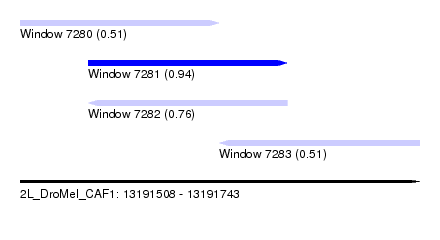
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,191,508 – 13,191,743 |
| Length | 235 |
| Max. P | 0.942218 |

| Location | 13,191,508 – 13,191,625 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.50 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13191508 117 + 22407834 UUUUCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUGUGUGAGCACAUAUGUACAUA-CAUA--UGCAUGUAAUGUGCAAGGUGCUGCAGCUUAAGAUGUUGCC ....(((((.....((((.(((((........)).)))))))((((((((((((((....)))))))))))))-)...--(((((.....))))))))))..(((((.......))))). ( -37.20) >DroSec_CAF1 11910 118 + 1 UUUUCGCCUCCUUUUUACCUUGGAAAAACUCAUCUGAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUA--UGUAUGUAAUGCGCAAGGUGUUGCAGCUUAAGAUGUUACC ........(((..........)))...((((((.....))))))(((.((((...(((((..((((((((.....)))--)))))(((((((.....))))))).)))))..))))))). ( -28.60) >DroSim_CAF1 11852 118 + 1 UUUUCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUA--UGUAUGUAUUGCGCAAGGUGUUGCAGCUUAAGAUGUUACC ........(((..........)))...((((((.....))))))(((.((((...(((((((((((((((.....)))--))))))).....((((....)))).)))))..))))))). ( -28.40) >DroYak_CAF1 12062 105 + 1 UUUUCGCCUCCUUUUUACCUUGGGAAAACUCAUCUCAAGUGAGUGUGUG--UGUGUGAGUACAUAUGUACGUA-CAUACGUGCAUGUAG------------UGCAGCUGAAGAUGUUGCC ..............(((((((((((.......)))))))...(((..((--((((((.((((....)))).))-))))))..)))))))------------.(((((.......))))). ( -32.10) >consensus UUUUCGCCUCCUUUUUACCUUGGAAAAACUCAUCUCAAGUGAGUGUGUGCGUAUGUGAGCACAUACAUACGUA_AAUA__UGCAUGUAAUGCGCAAGGUGUUGCAGCUUAAGAUGUUACC ........(((..........)))......(((((.((((..((((((((........))))))))..................((((..((((...)))))))))))).)))))..... (-19.88 = -19.50 + -0.38)

| Location | 13,191,548 – 13,191,665 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13191548 117 + 22407834 GAGUGUGUGCGUGUGUGAGCACAUAUGUACAUA-CAUA--UGCAUGUAAUGUGCAAGGUGCUGCAGCUUAAGAUGUUGCCAAGGGCAACGAUUCGCGGCUCGGAUUCGGAUGUAAAUCGA ..((((((((((((((....)))))))))))))-)...--(((((.....)))))..((.(((((((.......)))))..)).))..(((((..((..(((....))).))..))))). ( -41.50) >DroSec_CAF1 11950 118 + 1 GAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUA--UGUAUGUAAUGCGCAAGGUGUUGCAGCUUAAGAUGUUACCAAGGGCAACGAUUUGAGGCUCUGAUUCGGAUUUAAAUCGA ...((.((((((...(((((..((((((((.....)))--)))))(((((((.....))))))).)))))..))).))))).......(((((((((..((((...))))))))))))). ( -31.10) >DroSim_CAF1 11892 112 + 1 GAGUGUGUGCGUAUGUGAGCAUAUACAUAUGUAAAAUA--UGUAUGUAUUGCGCAAGGUGUUGCAGCUUAAGAUGUUACCAAGGGCAACGAUUUGCGGCUCUGA------UGUAAAUCGA ((.....(((((....((((((((((((((.....)))--)))))))....((((((.((((((..(((...........))).)))))).))))))))))..)------))))..)).. ( -34.60) >DroYak_CAF1 12102 105 + 1 GAGUGUGUG--UGUGUGAGUACAUAUGUACGUA-CAUACGUGCAUGUAG------------UGCAGCUGAAGAUGUUGCCAAGGGCAACAAUUUGAGGCUCUGAUUCGGAUGCUGAUCGG ..(((..((--((((((.((((....)))).))-))))))..)))....------------...((((..((((((((((...)))))).))))..))))(((((.(((...)))))))) ( -37.40) >consensus GAGUGUGUGCGUAUGUGAGCACAUACAUACGUA_AAUA__UGCAUGUAAUGCGCAAGGUGUUGCAGCUUAAGAUGUUACCAAGGGCAACGAUUUGAGGCUCUGAUUCGGAUGUAAAUCGA ..((((((((........))))))))..................((((..((((...))))))))(((..((((((((((...)))))).))))..)))..................... (-19.24 = -19.55 + 0.31)

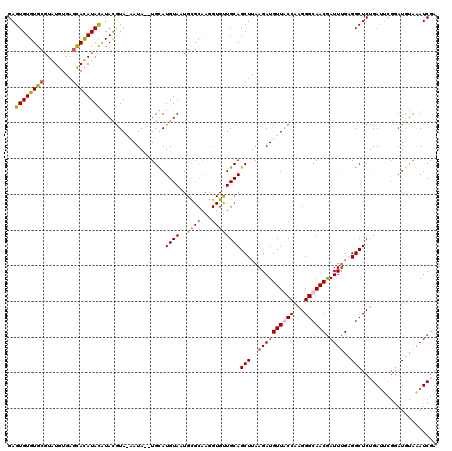
| Location | 13,191,548 – 13,191,665 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -11.71 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13191548 117 - 22407834 UCGAUUUACAUCCGAAUCCGAGCCGCGAAUCGUUGCCCUUGGCAACAUCUUAAGCUGCAGCACCUUGCACAUUACAUGCA--UAUG-UAUGUACAUAUGUGCUCACACACGCACACACUC (((((((......)))).)))...(((....((((((...)))))).......((....)).....(((((((((((((.--...)-))))))...)))))).......)))........ ( -30.20) >DroSec_CAF1 11950 118 - 1 UCGAUUUAAAUCCGAAUCAGAGCCUCAAAUCGUUGCCCUUGGUAACAUCUUAAGCUGCAACACCUUGCGCAUUACAUACA--UAUUUUACAUAUGUAUAUGCUCACAUACGCACACACUC ..(((((......))))).((((........((((((...)))))).......((.((((....)))))).....(((((--(((.....))))))))..))))................ ( -24.50) >DroSim_CAF1 11892 112 - 1 UCGAUUUACA------UCAGAGCCGCAAAUCGUUGCCCUUGGUAACAUCUUAAGCUGCAACACCUUGCGCAAUACAUACA--UAUUUUACAUAUGUAUAUGCUCACAUACGCACACACUC ..........------...(((((((((...(((((.(((((.......)))))..)))))...)))))......(((((--(((.....))))))))..))))................ ( -23.30) >DroYak_CAF1 12102 105 - 1 CCGAUCAGCAUCCGAAUCAGAGCCUCAAAUUGUUGCCCUUGGCAACAUCUUCAGCUGCA------------CUACAUGCACGUAUG-UACGUACAUAUGUACUCACACA--CACACACUC .......((((......(((...((.((..(((((((...)))))))..)).)))))..------------....))))(((((((-(....)))))))).........--......... ( -21.30) >consensus UCGAUUUACAUCCGAAUCAGAGCCGCAAAUCGUUGCCCUUGGCAACAUCUUAAGCUGCAACACCUUGCGCAUUACAUACA__UAUG_UACAUACAUAUAUGCUCACACACGCACACACUC ...................((((........((((((...))))))..........((((....))))................................))))................ (-11.71 = -12.40 + 0.69)

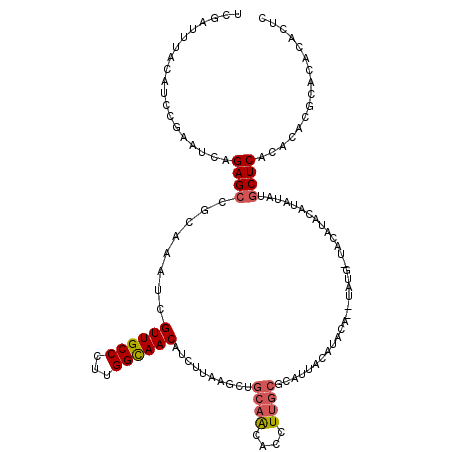
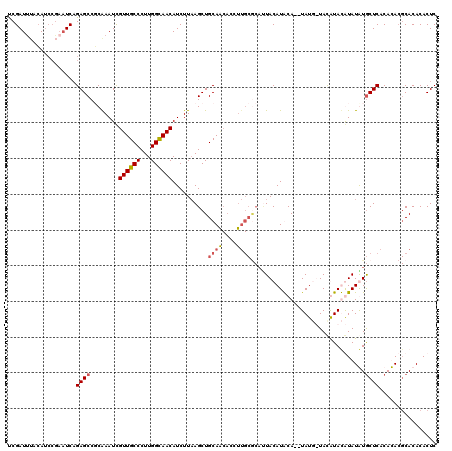
| Location | 13,191,625 – 13,191,743 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -18.58 |
| Energy contribution | -20.64 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13191625 118 - 22407834 ACAAAGGGUGAAAAAAAUGCGAAACCAGAAGUUUCGGUGGCUUC-AUAAUUACCUGGAAUGAGAUA-CUCCCGUUAUUUCUCGAUUUACAUCCGAAUCCGAGCCGCGAAUCGUUGCCCUU ...(((((..(........((((((.....))))))(((((((.-...............((((.(-(....))....))))(((((......))))).)))))))......)..))))) ( -30.90) >DroSec_CAF1 12028 109 - 1 ACAAAGGGUGAAA-AAAUGCGAAACCAGAAGUUUCGGUUAUUUCCGUUAU-UUCUCGAUU---------UCCGUUAUUUCUCGAUUUAAAUCCGAAUCAGAGCCUCAAAUCGUUGCCCUU ...((((((((((-.((((.(((((.((((((..(((......)))..))-)))).).))---------)))))).))))..(((((......)))))................)))))) ( -27.60) >DroSim_CAF1 11970 110 - 1 ACAAAGGGUGAAA-AAGUGCGAAACCAGAAGUUUCGGUGGCUUC-AUAAU-ACCUGGAAUUAGAUA-CUCCCGUUAUUUCUCGAUUUACA------UCAGAGCCGCAAAUCGUUGCCCUU ...(((((..(..-...((((.....((((((..(((..(..((-.((((-.......))))))..-)..)))..)))))).(((....)------)).....)))).....)..))))) ( -28.40) >DroYak_CAF1 12167 118 - 1 ACAAAGGGUGAAAAAAAAGCGAAACCAGAAGUUUCGGUGGCUUC-AUAAU-ACCAAGAACUAGAUAACCCCUGUUAUUACCCGAUCAGCAUCCGAAUCAGAGCCUCAAAUUGUUGCCCUU ...(((((..(........((((((.....))))))(.(((((.-.....-...........((((((....))))))....(((..(....)..))).))))).)......)..))))) ( -24.80) >consensus ACAAAGGGUGAAA_AAAUGCGAAACCAGAAGUUUCGGUGGCUUC_AUAAU_ACCUGGAAUUAGAUA_CUCCCGUUAUUUCUCGAUUUACAUCCGAAUCAGAGCCGCAAAUCGUUGCCCUU ...(((((..(........((((((.....))))))(((((((............((.............))..........(((((......))))).)))))))......)..))))) (-18.58 = -20.64 + 2.06)

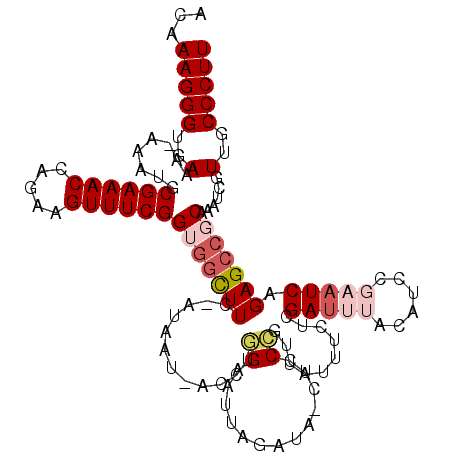
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:35 2006