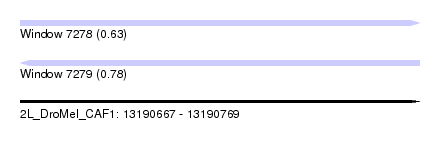
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,190,667 – 13,190,769 |
| Length | 102 |
| Max. P | 0.775946 |

| Location | 13,190,667 – 13,190,769 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13190667 102 + 22407834 AUAGAAUAUGGAAGGCGUAAAGCUUUUCUAUUUGUUGAU--AUUGACAACGGUUGGU---GGCCA---GCU---------GUUGGGCCAAAGCUGAUAAGAGCA-AUGAGCCCAUAAAAC ...(((((.(((((((.....))))))))))))(((...--...)))(((((((((.---..)))---)))---------)))((((((..(((......))).-.)).))))....... ( -32.30) >DroSec_CAF1 11063 120 + 1 AUAGAAUAUGGAAGGCGUAAAGCUUUUCUAUUUGUUGAUAAAUUGACAACGGUUGGUGGUGGCCAAAAGCUUGGGGAAAGGUUGGGGAAAAGCUGAUAAGAGCAAAUGAGCCCAUAAAAC ......(((((.........(((((((((..((((..(....)..))))...(((((....))))).(((((......)))))..))))))))).......((......))))))).... ( -25.90) >DroSim_CAF1 11024 108 + 1 AUAGAAUAUGGAAGGCGUAAAGCUUUUCUAUUUGUUGAUAAAUUGACAACGGUUGGUGGUGGCCA---GCU---------GUUGGGGAAAAGCUGAUAAGAACAAAUGAGCCCAUAAAAC ......((((((((((.....)))))..((((((((......((..(((((((((((....))))---)))---------))))..))....((....))))))))))...))))).... ( -30.30) >DroYak_CAF1 11200 104 + 1 AUGGAAUAUGGAAGGCGUAAAGCUUUUCUAUUUGUUGAUAAAUUGACAACGGUUAGU---GGCCA---GCU---------GUUGG-GAAAAGCUGAUAAGAGCAAAUGAGCCCAUAAAAC ((((.....(((((((.....)))))))((((((((.....((((((....))))))---...((---(((---------.....-....))))).....))))))))...))))..... ( -25.50) >consensus AUAGAAUAUGGAAGGCGUAAAGCUUUUCUAUUUGUUGAUAAAUUGACAACGGUUGGU___GGCCA___GCU_________GUUGGGGAAAAGCUGAUAAGAGCAAAUGAGCCCAUAAAAC ......(((((.........(((((((((..((((..(....)..)))).((((......)))).....................))))))))).......((......))))))).... (-18.88 = -19.88 + 1.00)

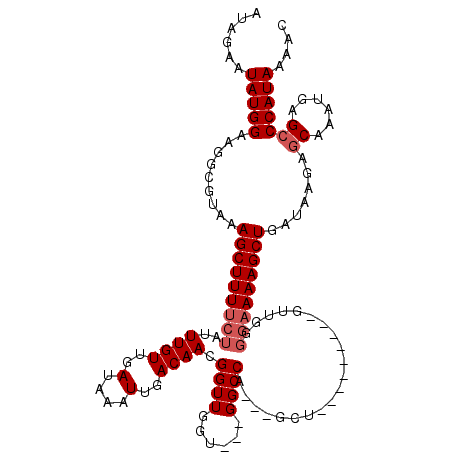

| Location | 13,190,667 – 13,190,769 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
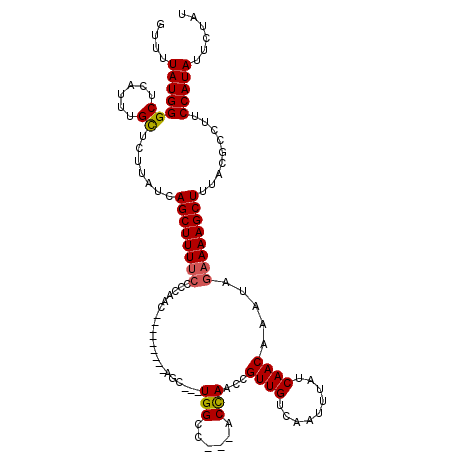
>2L_DroMel_CAF1 13190667 102 - 22407834 GUUUUAUGGGCUCAU-UGCUCUUAUCAGCUUUGGCCCAAC---------AGC---UGGCC---ACCAACCGUUGUCAAU--AUCAACAAAUAGAAAAGCUUUACGCCUUCCAUAUUCUAU (((((.(((((.((.-.(((......)))..)))))))((---------(((---.((..---.....)))))))....--.............)))))..................... ( -20.80) >DroSec_CAF1 11063 120 - 1 GUUUUAUGGGCUCAUUUGCUCUUAUCAGCUUUUCCCCAACCUUUCCCCAAGCUUUUGGCCACCACCAACCGUUGUCAAUUUAUCAACAAAUAGAAAAGCUUUACGCCUUCCAUAUUCUAU ....(((((((......)).......((((((((....................((((......))))..((((.........)))).....)))))))).........)))))...... ( -19.60) >DroSim_CAF1 11024 108 - 1 GUUUUAUGGGCUCAUUUGUUCUUAUCAGCUUUUCCCCAAC---------AGC---UGGCCACCACCAACCGUUGUCAAUUUAUCAACAAAUAGAAAAGCUUUACGCCUUCCAUAUUCUAU ....(((((((......)).......((((((((..((((---------.(.---(((......))).).))))...((((......)))).)))))))).........)))))...... ( -17.00) >DroYak_CAF1 11200 104 - 1 GUUUUAUGGGCUCAUUUGCUCUUAUCAGCUUUUC-CCAAC---------AGC---UGGCC---ACUAACCGUUGUCAAUUUAUCAACAAAUAGAAAAGCUUUACGCCUUCCAUAUUCCAU ....(((((((......)).......((((((((-(((..---------...---)))..---.......((((.........)))).....)))))))).........)))))...... ( -18.40) >consensus GUUUUAUGGGCUCAUUUGCUCUUAUCAGCUUUUCCCCAAC_________AGC___UGGCC___ACCAACCGUUGUCAAUUUAUCAACAAAUAGAAAAGCUUUACGCCUUCCAUAUUCUAU ....(((((((......)).......((((((((.....................(((......)))...((((.........)))).....)))))))).........)))))...... (-16.23 = -16.35 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:31 2006