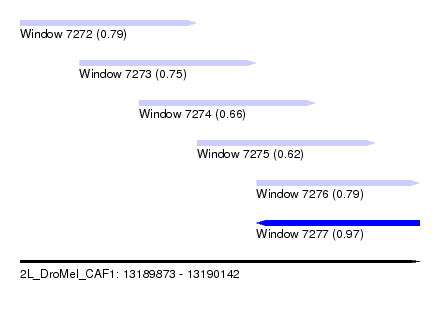
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,189,873 – 13,190,142 |
| Length | 269 |
| Max. P | 0.974130 |

| Location | 13,189,873 – 13,189,992 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.57 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13189873 119 + 22407834 GGCAACGGACAAAAUCAAUAAGUUAGGUAAAUAACAUAAAUAAAACGGUGGUCUAAAGGAGGCGGAGGACGCGACGUCCAGUUGCGCGUCCAAAUGGCGACGCACAGA-AAUGUUUGGAA .(((((((((...........((((......))))..........................(((.....)))...)))).)))))(((((........)))))(((..-..)))...... ( -29.90) >DroSec_CAF1 10268 119 + 1 GGCAAUGGACAAAAUCAAUAAGUUAGGUAAAUAAUAUAAAUAAAACGGUGGUCUAAAGGAGGCGGAGGACGCGCAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAA .((..(((((...(((......(((.(((.....)))...)))...))).)))))...........((((((((((.....)))))))))).....))..((((((..-..))))))... ( -33.00) >DroSim_CAF1 10249 119 + 1 GGCAAUGGACAAAAUCAAUAAGUUAGGUAAAUAAUAUAAAUAAAACGGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAA .((..((((((....((((...............................((((.....))))...((((.....)))).))))..))))))....))..((((((..-..))))))... ( -31.10) >DroYak_CAF1 10383 120 + 1 UGGAAUGAACAAAAUCAAUAAGUUAGGUAAAUAAUAUAAAUAAAACAGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAUUGGCGACGCACAGAAAAUGUGUGGAA ..............((...............................((.(((....(((.(((.(((((.....))))...).))).)))....))).))(((((.....))))).)). ( -25.30) >consensus GGCAAUGGACAAAAUCAAUAAGUUAGGUAAAUAAUAUAAAUAAAACGGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA_AAUGUGUGGAA .....................((((......))))............((.(((....(((.(((.(((((.....))))...).))).)))....))).))(((((.....))))).... (-24.70 = -24.57 + -0.13)

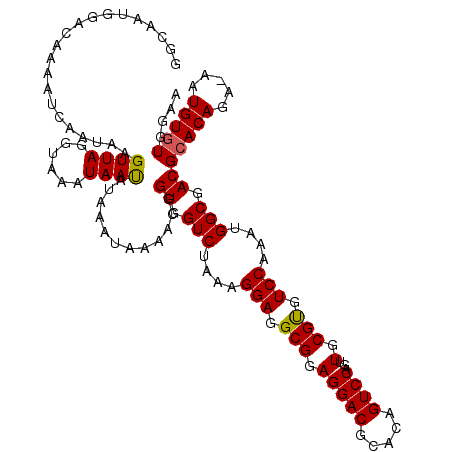
| Location | 13,189,913 – 13,190,032 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13189913 119 + 22407834 UAAAACGGUGGUCUAAAGGAGGCGGAGGACGCGACGUCCAGUUGCGCGUCCAAAUGGCGACGCACAGA-AAUGUUUGGAACCGUAAAGAGGUGGAACUUUUGUAGUAUUCUUCUUUUAAU ....(((((..(((((.....(((((.(.((((((.....))))))).)))....(....)))(((..-..)))))))))))))(((((((....(((.....)))...))))))).... ( -34.50) >DroSec_CAF1 10308 116 + 1 UAAAACGGUGGUCUAAAGGAGGCGGAGGACGCGCAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAU .............((((((((((...((((((((((.....))))))))))..((((...((((((..-..))))))...))))(((((......)))))....)).)))---))))).. ( -40.20) >DroSim_CAF1 10289 116 + 1 UAAAACGGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAU ....(((((.(((....(((.(((.(((((.....))))...).))).)))....)))..((((((..-..))))))..)))))....((((((((((.....))..)))---))))).. ( -36.30) >DroYak_CAF1 10423 119 + 1 UAAAACAGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAUUGGCGACGCACAGAAAAUGUGUGGAACCAUAAAGUGAGGGAACUUUUGUAGUAUUCUUC-UUAAAU .......(((((.....(((.(((.(((((.....))))...).))).))).........((((((.....))))))..))))).....(((((((((.....))).))))))-...... ( -32.80) >consensus UAAAACGGUGGUCUAAAGGAGGCGGAGGACGCACAGUCCAGUUGCGUGUCCAAAUGGCGACGCACAGA_AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC___UUUUAAU ....((((.(((((...(((.(((.(((((.....))))...).))).)))..((((...((((((.....))))))...))))........)).))).))))................. (-29.06 = -29.00 + -0.06)

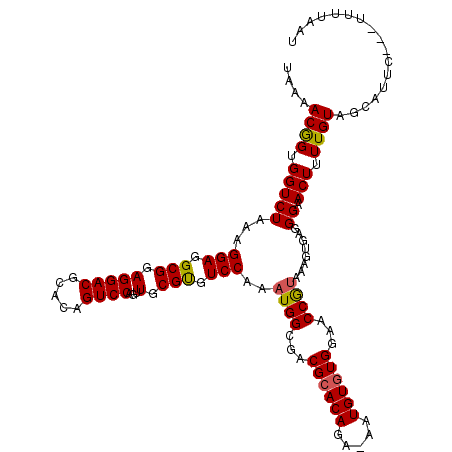
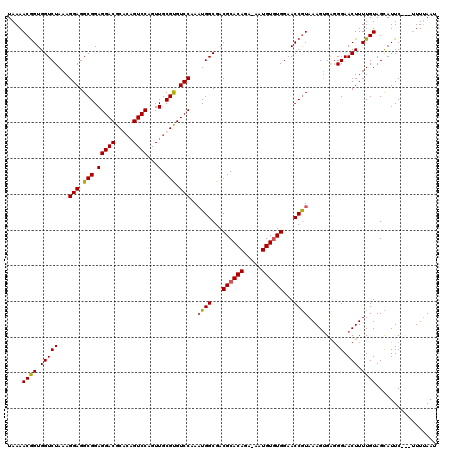
| Location | 13,189,953 – 13,190,072 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13189953 119 + 22407834 GUUGCGCGUCCAAAUGGCGACGCACAGA-AAUGUUUGGAACCGUAAAGAGGUGGAACUUUUGUAGUAUUCUUCUUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACAAGUUC (((.((((((((...(((((((((((((-(..((((...(((.......))).)))))))))).(((((.((.....)).))))).........))))).))))))))))))))...... ( -30.60) >DroSec_CAF1 10348 115 + 1 GUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAUAAUGCAUUAUAA-AAUGUCUGUCUGGAUGUGGACGAGUUU (((.((..((((.((((...((((((..-..))))))...)))).....((.(((..((((((((((((.---.......)))))..)))))-))..))).))))))..)))))...... ( -34.80) >DroSim_CAF1 10329 116 + 1 GUUGCGUGUCCAAAUGGCGACGCACAGA-AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUU (((.((..((((.((((...((((((..-..))))))...)))).....((.((((((.((((((((((.---.......)))))..))))).))).))).))))))..)))))...... ( -35.10) >DroYak_CAF1 10463 119 + 1 GUUGCGUGUCCAAUUGGCGACGCACAGAAAAUGUGUGGAACCAUAAAGUGAGGGAACUUUUGUAGUAUUCUUC-UUAAAUAAUGCAUUAUGAUGGUGUCUGUCUGGAUGUGAAGGAGUUG ....((..((((((.(((..((((((.....))))))..(((((.....(((((((((.....))).))))))-....((((....)))).)))))))).)).))))..))......... ( -28.10) >consensus GUUGCGUGUCCAAAUGGCGACGCACAGA_AAUGUGUGGAACCGUAAAGUGAGGGAACUUUUGUAGCAUUC___UUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUU (((.((((((((.((((...((((((.....))))))...)))).....((.((((((.((((((((((...........)))))..))))).))).))).)))))))))))))...... (-25.90 = -25.52 + -0.37)


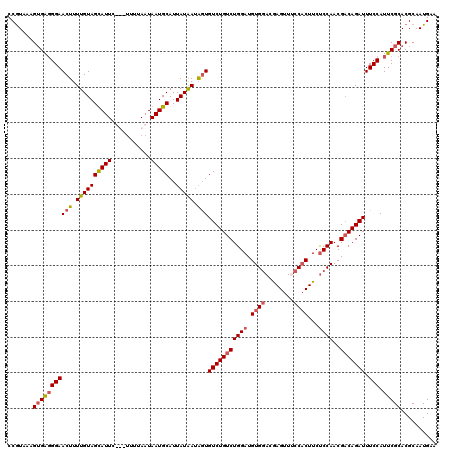
| Location | 13,189,992 – 13,190,112 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13189992 120 + 22407834 CCGUAAAGAGGUGGAACUUUUGUAGUAUUCUUCUUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACAAGUUCCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAA .(((...(((.(((((((.((((((((((.((.....)).)))))..))))).)))(((((((((((.((((........))))...))))..))))))).)))))))..)))....... ( -29.10) >DroSec_CAF1 10387 116 + 1 CCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAUAAUGCAUUAUAA-AAUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAA .......(((((((((.((((((((((((.---.......)))))..)))))-)).(((((((((((.(((((......)))))...))))..))))))))))).))))).......... ( -33.50) >DroSim_CAF1 10368 117 + 1 CCGUAAAGUGAGGGAACUUUUGUAGCAUUC---UUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAA .......(((((((((((.((((((((((.---.......)))))..))))).)))(((((((((((.(((((......)))))...))))..))))))).))).))))).......... ( -34.20) >DroYak_CAF1 10503 119 + 1 CCAUAAAGUGAGGGAACUUUUGUAGUAUUCUUC-UUAAAUAAUGCAUUAUGAUGGUGUCUGUCUGGAUGUGAAGGAGUUGCCUCUUCCCCAACGCCAGAUUUCCAUUCGCACGCAAUGAA .(((.....(((((((((.....))).))))))-........(((.....(((((.....((((((...((..((((......))))..))...))))))..))))).....)))))).. ( -26.80) >consensus CCGUAAAGUGAGGGAACUUUUGUAGCAUUC___UUUUAAUAAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAA .......(((((((((((.((((((((((...........)))))..))))).)))(((((((((((.((((........))))...))))..))))))).))).))))).......... (-22.91 = -23.85 + 0.94)

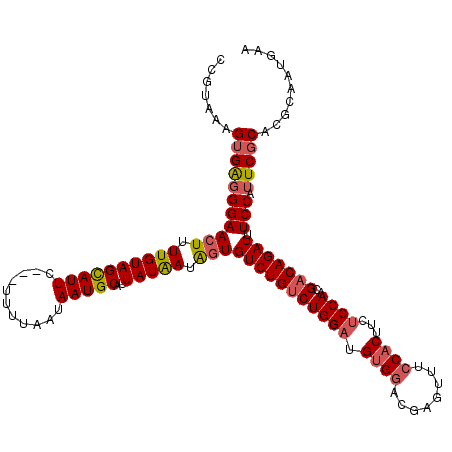
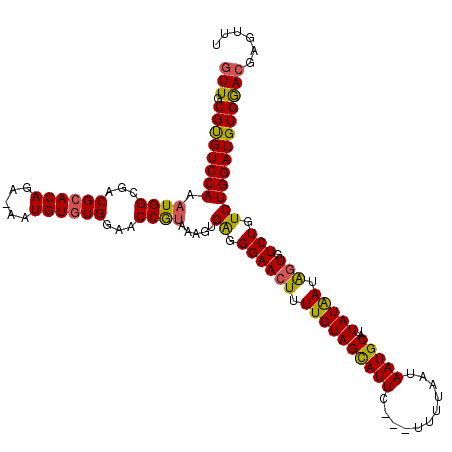
| Location | 13,190,032 – 13,190,142 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
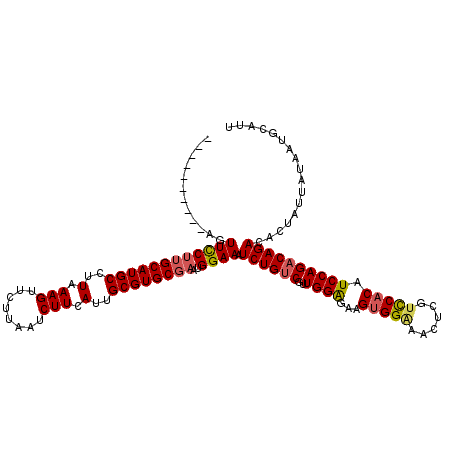
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -23.23 |
| Energy contribution | -24.48 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13190032 110 + 22407834 AAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACAAGUUCCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAAGAUUAAGAACUUUAAGGCAUGCAAGGAACU---------- ................(((((((((((.((((........))))...))))..)))))))((((.((.(((.((..(((((........)))))..)).)))))))))..---------- ( -29.60) >DroSec_CAF1 10424 109 + 1 AAUGCAUUAUAA-AAUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAAGAUUAAGAACUUUAAGGCAUGCAAGGAACU---------- ............-...(((((((((((.(((((......)))))...))))..)))))))((((.((.(((.((..(((((........)))))..)).)))))))))..---------- ( -30.90) >DroSim_CAF1 10405 110 + 1 AAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAAGAUUAAGAACUUUAAGGCAUGCAAGAAACU---------- ..(((((.........(((((((((((.(((((......)))))...))))..)))))))............((..(((((........)))))..))))))).......---------- ( -28.80) >DroYak_CAF1 10542 120 + 1 AAUGCAUUAUGAUGGUGUCUGUCUGGAUGUGAAGGAGUUGCCUCUUCCCCAACGCCAGAUUUCCAUUCGCACGCAAUGAAGAUUAAGAACUUUAGGGCAUGCUGGGAACUACAAUAUGUG ...((((..((.((((....((((((...((..((((......))))..))...))))))(((((...(((.((..(((((........)))))..)).))))))))))))))..)))). ( -31.40) >consensus AAUGCAUUAUAAUAGUGUCUGUCUGGAUGUGGACGAGUUUCCACUUCUCCAACGACAGAUUUCCAUUCGCACGCAAUGAAGAUUAAGAACUUUAAGGCAUGCAAGGAACU__________ ................(((((((((((.((((........))))...))))..)))))))((((....(((.((..(((((........)))))..)).)))..))))............ (-23.23 = -24.48 + 1.25)

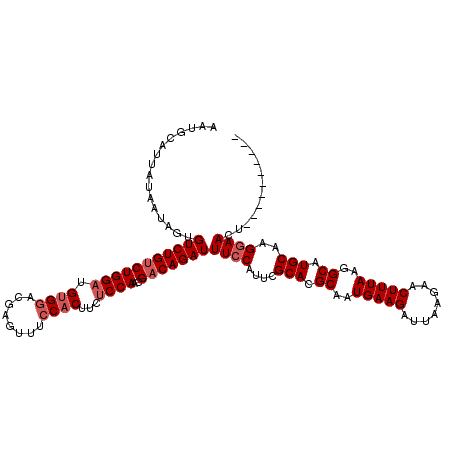

| Location | 13,190,032 – 13,190,142 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13190032 110 - 22407834 ----------AGUUCCUUGCAUGCCUUAAAGUUCUUAAUCUUCAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGGAACUUGUCCACAUCCAGACAGACACUAUUAUAAUGCAUU ----------..((((((((((((..(.(((........))).)..))))))))..)))).((((((..((((...(((((......))))).))))))))))................. ( -32.10) >DroSec_CAF1 10424 109 - 1 ----------AGUUCCUUGCAUGCCUUAAAGUUCUUAAUCUUCAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACAUU-UUAUAAUGCAUU ----------..((((((((((((..(.(((........))).)..))))))))..)))).((((((..((((...(((((......))))).))))))))))((((-....)))).... ( -33.20) >DroSim_CAF1 10405 110 - 1 ----------AGUUUCUUGCAUGCCUUAAAGUUCUUAAUCUUCAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACACUAUUAUAAUGCAUU ----------.......(((((((..(.(((........))).)..)))))))(((((...((((((..((((...(((((......))))).))))))))))..))))).......... ( -30.80) >DroYak_CAF1 10542 120 - 1 CACAUAUUGUAGUUCCCAGCAUGCCCUAAAGUUCUUAAUCUUCAUUGCGUGCGAAUGGAAAUCUGGCGUUGGGGAAGAGGCAACUCCUUCACAUCCAGACAGACACCAUCAUAAUGCAUU .......((((.......((((((....(((........)))....))))))((.(((...((((...(((((((((((....)).))))...))))).))))..)))))....)))).. ( -30.10) >consensus __________AGUUCCUUGCAUGCCUUAAAGUUCUUAAUCUUCAUUGCGUGCGAAUGGAAAUCUGUCGUUGGAGAAGUGGAAACUCGUCCACAUCCAGACAGACACUAUUAUAAUGCAUU ............((((((((((((..(.(((........))).)..))))))))..)))).((((((..((((...(((((......))))).))))))))))................. (-27.04 = -27.60 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:29 2006