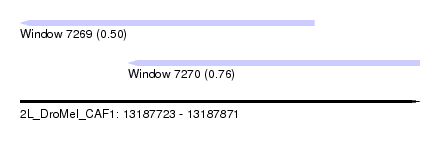
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,187,723 – 13,187,871 |
| Length | 148 |
| Max. P | 0.759984 |

| Location | 13,187,723 – 13,187,832 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.65 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -22.27 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
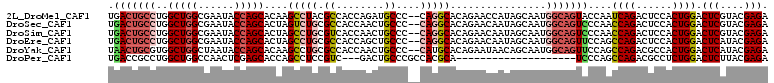
>2L_DroMel_CAF1 13187723 109 - 22407834 ACCAGAUGCCC--CAGGCACAGAACCAUAGCAAUGGCAGUACCAAUCAGACUCCACUGGACUCGUACGAGAGUAUCGAUCAUCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ......((((.--..)))).....(((((....(((((((((...((((......))))....))))(((.((((((((......)))))))).)))...))))).))))) ( -32.10) >DroPse_CAF1 9838 88 - 1 ---GACUGCCCGCCACGCA--------------------UCCCAGCCAGACGCCUCUGGACUCUUACGAGAGCAUUGAUGACCAGAUCGAUGCGUUGAAGUGCCAUUAUGG ---......(((....(((--------------------(..((((((((....))))..(((....))).((((((((......))))))))))))..)))).....))) ( -22.90) >DroSec_CAF1 8147 109 - 1 ACCAACUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGAGUAUCGACCAUCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ....((((((.--...((...........))...)))))).(((.((((......))))(((..((((((.(((((((........))))))).)))..)))..))).))) ( -31.10) >DroSim_CAF1 8128 109 - 1 ACCAACUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGAGUAUCGACCAUCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ....((((((.--...((...........))...)))))).(((.((((......))))(((..((((((.(((((((........))))))).)))..)))..))).))) ( -31.10) >DroEre_CAF1 8293 109 - 1 ACCAGCUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUUCCAGCCAGACUCCACUGGACUCAUACGAGAGUAUCGACCAUCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ...(((((((.--...((...........))...)))))))(((.((((......))))(((.(((((((.(((((((........))))))).)))..)))).))).))) ( -32.50) >DroYak_CAF1 8231 109 - 1 ACCAACUGCCC--CAUGCACAGAAUAACAGCAAUGGCAGUUCCAGCCAGACGCCACUGGACUCAUACGAGAGUAUCGACCACCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ...(((((((.--..(((...........)))..)))))))(((.((((......))))(((.(((((((.(((((((........))))))).)))..)))).))).))) ( -33.20) >consensus ACCAACUGCCC__CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGAGUAUCGACCAUCAAAUCGAUGCGCUCAAGUGUCAGUAUGG ....((((((......((...........))...)))))).(((.((((......))))(((..((((((.(((((((........))))))).)))..)))..))).))) (-22.27 = -23.17 + 0.89)
| Location | 13,187,763 – 13,187,871 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -25.16 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
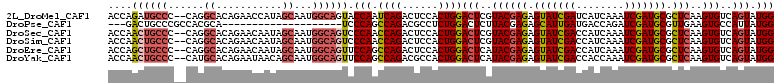
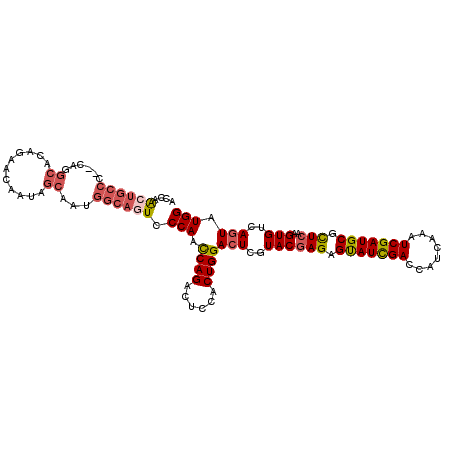
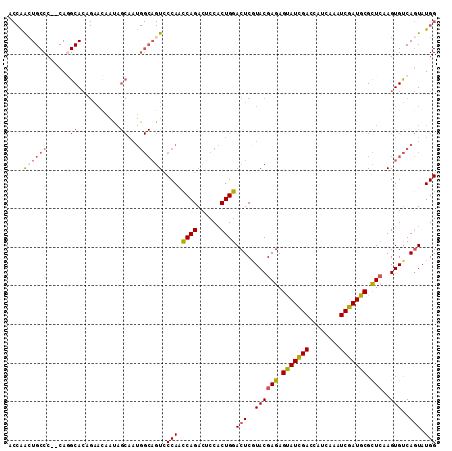
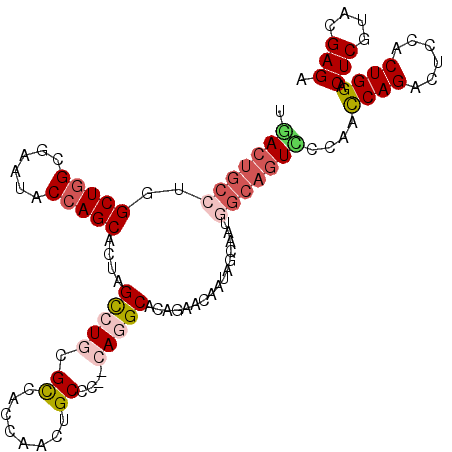
>2L_DroMel_CAF1 13187763 108 - 22407834 UGACUGCCUGGCUGGCGAAUACCAGCACAAGCCUACGCCACCAGAUGCCC--CAGGCACAGAACCAUAGCAAUGGCAGUACCAAUCAGACUCCACUGGACUCGUACGAGA ..(((((((((((((......))))).)).((((..((........))..--.))))................)))))).....((((......)))).(((....))). ( -29.70) >DroSec_CAF1 8187 108 - 1 UGACUGCCUGGCUGGCGAAUACCAGCACUAGUCUGCGCCACCAACUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGA .(((((((..(((((......))))).(((.((((.(((...........--..))).))))....)))....)))))))....((((......)))).(((....))). ( -38.72) >DroSim_CAF1 8168 108 - 1 UGACUGCCUGGCUGGCGAAUACCAGCACUAGCCUGCGUCACCAACUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGA .(((((((..(((((......)))))....(((((.(.((.....)).).--)))))................)))))))....((((......)))).(((....))). ( -37.10) >DroEre_CAF1 8333 108 - 1 UGACUGCCUGGCUGGCGAAUACCAGCACUAGCCUGCGCCACCAGCUGCCC--CAGGCACAGAACAAUAGCAAUGGCAGUUCCAGCCAGACUCCACUGGACUCAUACGAGA .(((((((..(((((......)))))....(((((.((........))..--)))))................)))))))....((((......)))).(((....))). ( -34.50) >DroYak_CAF1 8271 108 - 1 UAACUGCGUGGCUGGCUAAUACCAGCACAAGCCUGCGCCACCAACUGCCC--CAUGCACAGAAUAACAGCAAUGGCAGUUCCAGCCAGACGCCACUGGACUCAUACGAGA .....((((((((.(((......)))...)))).))))....(((((((.--..(((...........)))..)))))))....((((......)))).(((....))). ( -32.50) >DroPer_CAF1 9999 87 - 1 UGACCGCCUGGCUGGCCAACUCGAGCACCAGCCUCCGUC---GACUGCCCGCCACGCA--------------------UCCCAGCCAGACGCCUCUGGACUCUUACGAGA .....((.((((.(((....((((((....)).....))---))..))).)))).)).--------------------......(((((....))))).(((....))). ( -25.40) >consensus UGACUGCCUGGCUGGCGAAUACCAGCACUAGCCUGCGCCACCAACUGCCC__CAGGCACAGAACAAUAGCAAUGGCAGUCCCAACCAGACUCCACUGGACUCGUACGAGA .(((((((..(((((......)))))....(((((.((........))....)))))................)))))))....((((......)))).(((....))). (-25.16 = -26.38 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:22 2006