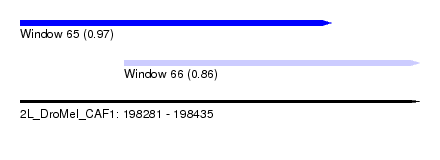
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 198,281 – 198,435 |
| Length | 154 |
| Max. P | 0.967144 |

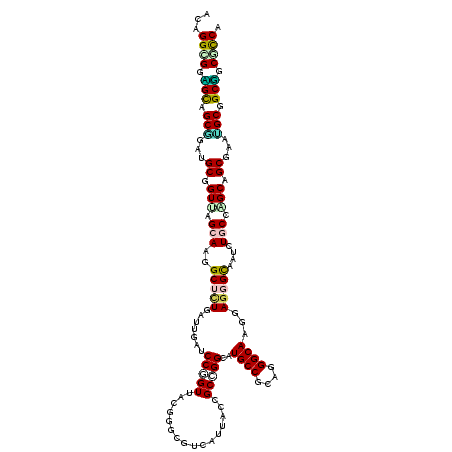
| Location | 198,281 – 198,401 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -54.93 |
| Consensus MFE | -33.52 |
| Energy contribution | -33.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 198281 120 + 22407834 ACCGGUGGGGUUGCGGAUGCGGUCAGCAAGGCUCUAGUUGAUCCAGUUACUGGAGUUAUCACCGCUGGAAUGCCCCAGGGCAAAGAGGGUAAUCUUCCAGCAGCUACCGCUGCUGCACCU ...((((.(((.((((..(((((.(....(((((((((..........))))))))).).)))(((((((((((....))))..((......))))))))).))..)))).))).)))). ( -51.40) >DroVir_CAF1 13837 120 + 1 ACAGGUGGUGCCGCGGAUGCAGUUAGCAAGGCUUUAAUUGAUCCCGUUACGGGUGUCAUUACUGCGGGCAUGCCGCAGGGCAAGGAGGGCAAUUUGCCGGCUGCGAAUGCGGCGGCGCCC ...((((.(((((((..((((((..((((.(((((..(((..((((...))))........((((((.....))))))..)))..)))))...))))..))))))..))))))).)))). ( -60.20) >DroGri_CAF1 14027 120 + 1 ACUGGCGGUGCAGCGGAUGCGGUCAGCAAGGCUCUGAUUGACCCGGUCACAGGCAUAAUUACGGCCGGAAUGCCGCAGGGCAAGGAAGGUAAUUUGCCGGCAGCGAAUGCGGCGGCGCCC ...((((.(((.(((..(((((((((((......)).))))))..(((...((((.((((((..((....((((....)))).))...))))))))))))).)))..))).))).)))). ( -47.10) >DroWil_CAF1 14741 120 + 1 ACUGGCGGAGCGGCAGAUGCUGUUAGCAAGGCAUUGGUGGAUCCUGUGACGGGUGUUAUAACCGCUGGCAUGCCACAGGGCAAAGAGGGUAAUCUGCCAGCAGCAACAGCUGCUGCUCCA ......(((((((((...((((((.((........(((.(((((((...)))).)))...)))((((((.((((....)))).(((......))))))))).))))))))))))))))). ( -54.30) >DroMoj_CAF1 13952 120 + 1 ACAGGCGGCGCCGCAGAUGCGGUGAGCAAGGCGCUGAUUGAUCCGGUUACGGGCGUCAUUACGGCGGGCAUGCCGCAGGGCAAGGAGGGCAAUUUGCCGGCUGCCAAUGCAGCGGCGCCA ......((((((((.((((((((.(((.....))).)))...(((....)))))))).....(((((.(.((((....)))).....(((.....)))).)))))......)))))))). ( -58.30) >DroAna_CAF1 13481 120 + 1 ACCGGCGGAGCAGCCGACGCGGUUAGCAAGGCUCUGGUGGAUCCUGUCACGGGCGUCAUCACAGCCGGCAUGCCUCAGGGCAAGGAGGGCAACCUCCCAGCAGCUACGGCCGCUGCUUCA ......((((((((....((.((.(((..((((.((((((..((((...))))..)))))).)))).((.((((....)))).(((((....)))))..)).))))).)).)))))))). ( -58.30) >consensus ACAGGCGGAGCAGCGGAUGCGGUUAGCAAGGCUCUGAUUGAUCCGGUUACGGGCGUCAUUACCGCCGGCAUGCCGCAGGGCAAGGAGGGCAAUCUGCCAGCAGCGAAUGCGGCGGCGCCA ...((((.(((.(((...((.(((.(((..(((((.......(((((................)))))..((((....))))...)))))....))).))).))...))).))).)))). (-33.52 = -33.44 + -0.08)

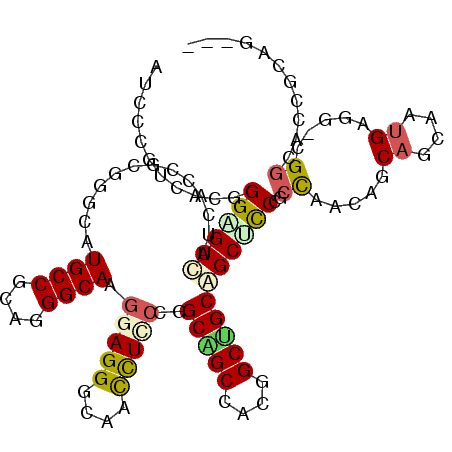
| Location | 198,321 – 198,435 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 198321 114 + 22407834 AUCCAGUUACUGGAGUUAUCACCGCUGGAAUGCCCCAGGGCAAAGAGGGUAAUCUUCCAGCAGCUACCGCUGCUGCACCUGCCAACAGCAGCAAUGAAGACGGUCAAGCAGCUC .(((((...)))))(((...((((((((((((((....))))..((......))))))))).......(((((((..........))))))).........)))..)))..... ( -38.80) >DroVir_CAF1 13877 106 + 1 AUCCCGUUACGGGUGUCAUUACUGCGGGCAUGCCGCAGGGCAAGGAGGGCAAUUUGCCGGCUGCGAAUGCGGCGGCGCCCGCCAACAGCAGCAAUGAAA-CGGUGCC------- ...(((((.((((((((....((((((.....))))))((((((........)))))).(((((....))))))))))))).......((....)).))-)))....------- ( -46.50) >DroPse_CAF1 15603 110 + 1 AUCCCGUCACGGGAGUUAUCACCGCGGGCAUGCCGCAGGGCAAGGAGGGCAACCUCCCGGCAGCCACGGCUGCAGCUCCCACCAACAGCAGCAAUGAGG-CGGCACCGCAG--- .(((((...))))).........((((...((((((.((((..(((((....)))))..(((((....))))).))))..........((....))..)-)))))))))..--- ( -52.50) >DroMoj_CAF1 13992 106 + 1 AUCCGGUUACGGGCGUCAUUACGGCGGGCAUGCCGCAGGGCAAGGAGGGCAAUUUGCCGGCUGCCAAUGCAGCGGCGCCAGCUAACAGCAGCAAUGAGG-UGACGCC------- ..(((....)))((((((((..(((.(((..(((....((((((........)))))).(((((....))))))))))).))).....((....)).))-)))))).------- ( -44.20) >DroAna_CAF1 13521 114 + 1 AUCCUGUCACGGGCGUCAUCACAGCCGGCAUGCCUCAGGGCAAGGAGGGCAACCUCCCAGCAGCUACGGCCGCUGCUUCAGCCAACAGCAGCAGCGACAUGGGACCACUGGGUC ..((((((..((.(((......(((..((.((((....)))).(((((....)))))..)).)))))).))((((((...((.....)))))))))))).))((((....)))) ( -49.20) >DroPer_CAF1 15789 110 + 1 AUCCCGUCACGGGAGUUAUCACCGCGGGCAUGCCGCAGGGCAAGGAGGGCAACCUCCCGGCAGCCACGGCUGCAGCUCCCACCAACAGCAGCAAUGAGG-CGGCACCGCAG--- .(((((...))))).........((((...((((((.((((..(((((....)))))..(((((....))))).))))..........((....))..)-)))))))))..--- ( -52.50) >consensus AUCCCGUCACGGGAGUCAUCACCGCGGGCAUGCCGCAGGGCAAGGAGGGCAACCUCCCGGCAGCCACGGCUGCAGCUCCCGCCAACAGCAGCAAUGAGG_CGGCACCGCAG___ ...........((((((.............((((....)))).(((((....)))))..(((((....)))))))))))................................... (-24.77 = -25.00 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:06 2006