| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,173,505 – 13,173,641 |
| Length | 136 |
| Max. P | 0.935718 |

| Location | 13,173,505 – 13,173,601 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13173505 96 + 22407834 CCCUCCUCGGUGCUGCCCGGAGCAGUGGCAUCGCCCACUGCUCCGCCGUUUAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCA .((......((((....(((((((((((......))))))))))).........((((((..((...))..)))))).....))))......)).. ( -35.90) >DroSec_CAF1 22268 96 + 1 CCCUCCUCGGUGAUGCCCGGAGCUGUGGCAUCGCCCACUGCUCCGCCGUUUAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCA ......(((((((((..((((((.((((......)))).)))))).........((((((..((...))..))))))........))))))))).. ( -32.60) >DroSim_CAF1 14631 96 + 1 CCCUCCUCGCUGAGGCCCGGAGCUGUGGCAUCGCCCACUGCUCCGCCGUUUAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCA ........((..((((..(((((.((((......)))).)))))))).......((((((..((...))..)))))).............)..)). ( -33.50) >DroEre_CAF1 14803 96 + 1 CCCUCUUCGGUGCUUCCCGGAGCUGUGGCAUCUCCCACUGCUCCGCCGUUCAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAGUGGCA .((.((...((((....((((((.((((......)))).)))))).........((((((..((...))..)))))).....))))...)).)).. ( -33.70) >DroWil_CAF1 78192 96 + 1 CCGUUUGCCUGGCCAGCUGGGGCGACAGCAUCGCCUACAUCGCCACCAUUCAAUGCGGCUAUCGUCUCAUCGGUGGCCAUUUGUAUUGUAUUGGCA .....((((((((......((((((.....)))))).....)))).....((((((((((((((......))))))))....))))))....)))) ( -35.30) >DroYak_CAF1 22738 96 + 1 CCCUCCUCGUUGCUUCCCGGAGCUGCGGCAUCUCCCACAGCUCCGCCGUUCAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCA .((.....((((.....((((((((.((......)).)))))))).....))))((((((..((...))..))))))...............)).. ( -31.20) >consensus CCCUCCUCGGUGCUGCCCGGAGCUGUGGCAUCGCCCACUGCUCCGCCGUUCAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCA ...........(((...((((((.((((......)))).))))))..((.(((.((((((..((...))..))))))...))).))......))). (-22.57 = -22.63 + 0.06)

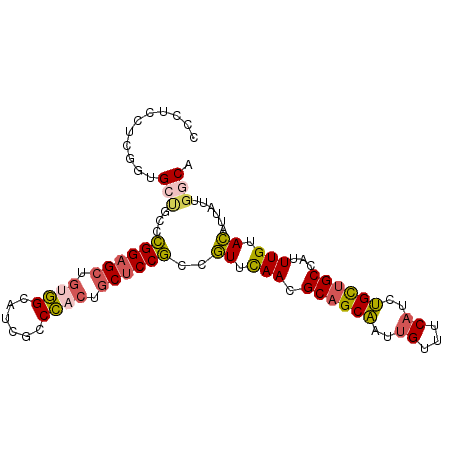

| Location | 13,173,505 – 13,173,601 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -25.55 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13173505 96 - 22407834 UGCCAAUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCGAUGCCACUGCUCCGGGCAGCACCGAGGAGGG ((((...............(((((((.((...)).))))))).........(((((((((((......)))))))))))))))...((.....)). ( -40.90) >DroSec_CAF1 22268 96 - 1 UGCCAAUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCGAUGCCACAGCUCCGGGCAUCACCGAGGAGGG ((((...............(((((((.((...)).))))))).........((((((.((((......)))).))))))))))...((.....)). ( -35.40) >DroSim_CAF1 14631 96 - 1 UGCCAAUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCGAUGCCACAGCUCCGGGCCUCAGCGAGGAGGG .(((...............(((((((.((...)).)))))))(.....))))(((((.((((......)))).)))))...((((......)))). ( -35.20) >DroEre_CAF1 14803 96 - 1 UGCCACUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUGAACGGCGGAGCAGUGGGAGAUGCCACAGCUCCGGGAAGCACCGAAGAGGG (((..((..(((.(((...(((((((.((...)).))))))).))).))).((((((.((((......)))).))))))))..)))((.....)). ( -33.80) >DroWil_CAF1 78192 96 - 1 UGCCAAUACAAUACAAAUGGCCACCGAUGAGACGAUAGCCGCAUUGAAUGGUGGCGAUGUAGGCGAUGCUGUCGCCCCAGCUGGCCAGGCAAACGG ((((.......((((..(.(((((((..((..((.....))..))...))))))).)))))(((.(.((((......))))).))).))))..... ( -31.90) >DroYak_CAF1 22738 96 - 1 UGCCAAUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUGAACGGCGGAGCUGUGGGAGAUGCCGCAGCUCCGGGAAGCAACGAGGAGGG ..((...............(((((((.((...)).)))))))((((.....(((((((((((......))))))))))).....)))).....)). ( -39.00) >consensus UGCCAAUAAUGUACAAAUGGCAGCAGAUGAAACAAUUGCUGCGUUAAACGGCGGAGCAGUGGGCGAUGCCACAGCUCCGGGCAGCACCGAGGAGGG ((((...............(((((((.((...)).))))))).........((((((.((((......)))).))))))))))............. (-25.55 = -25.80 + 0.25)
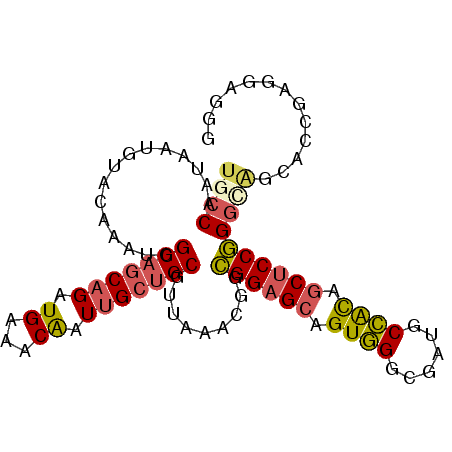
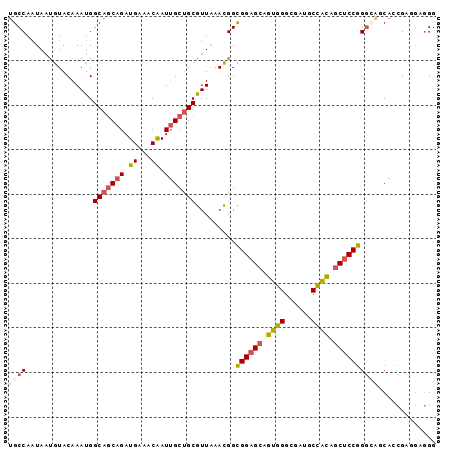
| Location | 13,173,521 – 13,173,641 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -46.15 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
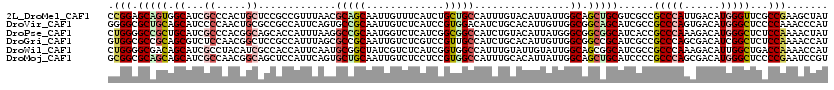
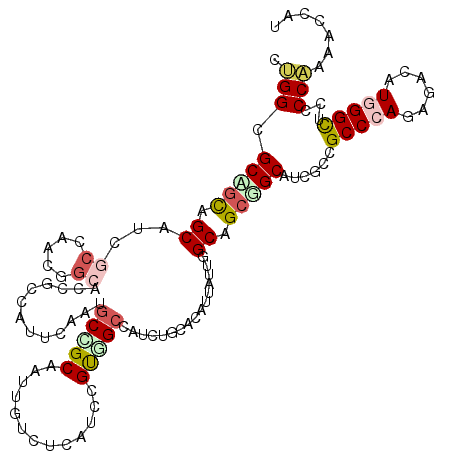
>2L_DroMel_CAF1 13173521 120 + 22407834 CCGGAGCAGUGGCAUCGCCCACUGCUCCGCCGUUUAACGCAGCAAUUGUUUCAUCUGCUGCCAUUUGUACAUUAUUGGCAGCUGCGUCGCCGCCCAUUGACAUGGGUUCGCCGAAGCUAU ..((((((((((......))))))))))((.((.....)).))....(((((....(((((((............))))))).(((.....((((((....)))))).))).)))))... ( -48.70) >DroVir_CAF1 17874 120 + 1 GGGGCGCUGCAGCAUCCCCAACUGCGCCGCCAUUCAGUGCCGCAAUUGUCUCAUCCGUGGACAUCUGCACAUUGUUGGCGGCAGCAUCGCCGCCCAGUGACAUGGGCUCCCCAAACCCAU ((((((.(((.(((........)))(((((((..(((((..(((..((((.((....))))))..))).))))).))))))).))).))))(((((......)))))...))........ ( -55.60) >DroPse_CAF1 44479 120 + 1 CUGGGGCCGCUGCAUCGCCCACGGCAGCACCAUUUAAGGCCGCAAUGGUCUCAUCGGCGGCCAUCUGUACAUUAUGGGCGGCGGCAUCACCGCCCAAAGACAUGGGCUCUCCAAAACUAU .(((((..(((((..((((((.((.....))......((((((.(((....)))..))))))............)))))))))))......(((((......))))).)))))....... ( -48.40) >DroGri_CAF1 46289 120 + 1 GUGGCGCCGCAGCGUCUCCAACGGCUCCGCCAUUUAGCGCCGCAAUUGUCUCGUCCGUUGCCAUCUGCACAUUGUUGGCGGCCGCAUCGCCGCCCAGCGACAUCGGCUCUCCAAAACCAU ((((.((((............)))).)))).....((.((((....((((......((.((.....))))...(((((((((......)))).))))))))).)))).)).......... ( -38.30) >DroWil_CAF1 78208 120 + 1 CUGGGGCGACAGCAUCGCCUACAUCGCCACCAUUCAAUGCGGCUAUCGUCUCAUCGGUGGCCAUUUGUAUUGUAUUGGCAGCGGCAUCGCCGCCCAAAGACAUUGGCUGACCAAAACCAU .((((((((.....)))))......(((((((..((((((((((((((......))))))))....))))))...)))..(((((...)))))..........))))...)))....... ( -43.80) >DroMoj_CAF1 59984 120 + 1 GCGGCGCAGCAGCAUCGCCAACGGCAGCUCCAUUCAGUGCUGCAAUUGUCUCCUCCGUGGCCAUUUGCACAUUAUUGGCAGCUGCAUCCCCGCCCAGCGACAUGGGCUCCCCGAAUCCGU (((..((....))..)))..((((((((((((...((((.(((((..(((........)))...)))))))))..))).))))))......(((((......)))))..........))) ( -42.10) >consensus CUGGCGCAGCAGCAUCGCCAACGGCACCGCCAUUCAAUGCCGCAAUUGUCUCAUCCGUGGCCAUCUGCACAUUAUUGGCAGCGGCAUCGCCGCCCAGAGACAUGGGCUCCCCAAAACCAU .(((.(((((.((...((.....)).............(((((.............)))))................)).)))))......(((((......)))))...)))....... (-21.73 = -22.40 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:10 2006