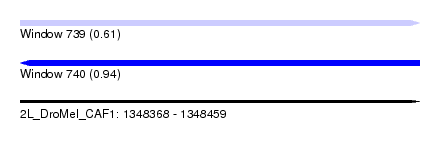
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,348,368 – 1,348,459 |
| Length | 91 |
| Max. P | 0.941775 |

| Location | 1,348,368 – 1,348,459 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
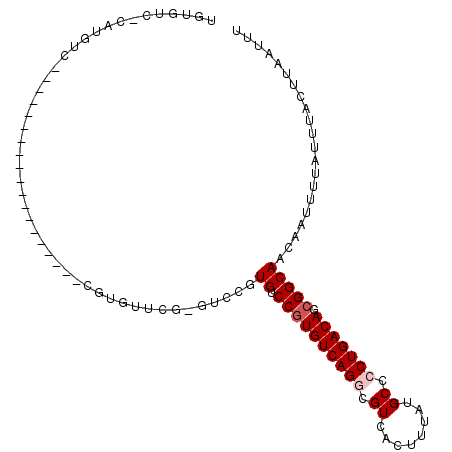
>2L_DroMel_CAF1 1348368 91 + 22407834 UGUGUCCCAUGUCCUGGC------------CGCAUGUUCU----CGUGUCCGUGUCAGGCGUCACUUUAUGCCCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU .(((............((------------((((((..(.----...)..)))((((((.((........)).)))))).)))))............)))....... ( -22.45) >DroPse_CAF1 148717 87 + 1 UGUGUC-CGUGUC-------------------CGUGUCGGAGUCCGUGUCCGUGUCAGACGUCACUUUAUGCGCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU (((.((-((....-------------------.....)))).....((.(((((((((.(((........))).)))))).)))))))).................. ( -20.30) >DroSec_CAF1 104898 91 + 1 UGUGUCCCAUGUCCUGGC------------CUCAUGUUCU----CGUGUCCGUGUCAGGCGUCACUUUAUGCCCCUGACAGGGGCAACAAUUUUAUUUACUUAAUUU .(((....(((.((((((------------(.((((....----))))...).))))))))))))....((((((.....))))))..................... ( -25.70) >DroWil_CAF1 148091 81 + 1 CGUGUC-CAUGUC-------------------------CAUGUCCAUGUCCGUGUCAGGCGUCACUUUAUGCGCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU ((((..-(((...-------------------------.)))..)))).(((((((((((((........)))))))))).)))....................... ( -26.80) >DroAna_CAF1 129882 106 + 1 -AUGUCCCAUGUCCUGCCUGGAAGUUUGUGCUCGUGUUCGUGUCCAUGUCCGUGUCAGGCGUCACUUUAUGCCCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU -........(((..((((.(((....((..(.((....)).)..))..))).(((((((.((........)).)))))))..))))))).................. ( -25.30) >DroPer_CAF1 157491 81 + 1 UGUGUC--------------------------CGUGUCGGAGUCCGUGUCCGUGUCAGACGUCACUUUAUGCGCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU (((..(--------------------------((((((((.((.((((...(((.(....).)))..)))).)))))))).)))..))).................. ( -20.30) >consensus UGUGUC_CAUGUC___________________CGUGUUCG_GUCCGUGUCCGUGUCAGGCGUCACUUUAUGCCCCUGACAGCGGCAACAAUUUUAUUUACUUAAUUU ..............................................((.((((((((((.((........)).))))))).)))))..................... (-14.38 = -14.88 + 0.50)

| Location | 1,348,368 – 1,348,459 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
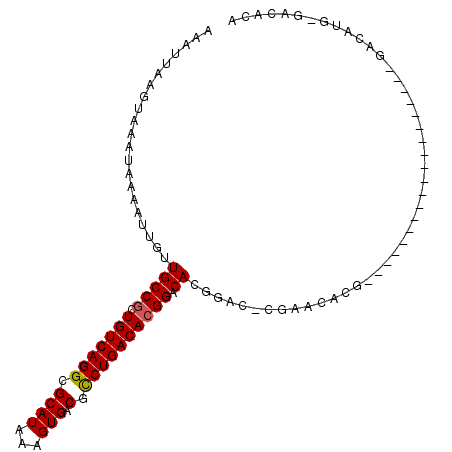
>2L_DroMel_CAF1 1348368 91 - 22407834 AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGGGCAUAAAGUGACGCCUGACACGGACACG----AGAACAUGCG------------GCCAGGACAUGGGACACA ..................((((((((((((((((.((((...))).).)))))))((....))----.......)))------------))...))))......... ( -25.00) >DroPse_CAF1 148717 87 - 1 AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGCGCAUAAAGUGACGUCUGACACGGACACGGACUCCGACACG-------------------GACACG-GACACA ..................((((.(((.((((((((((((...))).))))))))))))...((...((((...))-------------------))..))-)))).. ( -26.10) >DroSec_CAF1 104898 91 - 1 AAAUUAAGUAAAUAAAAUUGUUGCCCCUGUCAGGGGCAUAAAGUGACGCCUGACACGGACACG----AGAACAUGAG------------GCCAGGACAUGGGACACA .......((........((((.(.((.(((((((.((((...))).).))))))).)).))))----).........------------.(((.....))).))... ( -20.90) >DroWil_CAF1 148091 81 - 1 AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGCGCAUAAAGUGACGCCUGACACGGACAUGGACAUG-------------------------GACAUG-GACACG ..................((((.(((.((((((((((((...))).)))))))))))).((((......-------------------------..))))-)))).. ( -28.00) >DroAna_CAF1 129882 106 - 1 AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGGGCAUAAAGUGACGCCUGACACGGACAUGGACACGAACACGAGCACAAACUUCCAGGCAGGACAUGGGACAU- ..................((((.(((.(((((((.((((...))).).)))))))))).((((..(.((....)).((............)).)..)))).)))).- ( -26.60) >DroPer_CAF1 157491 81 - 1 AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGCGCAUAAAGUGACGUCUGACACGGACACGGACUCCGACACG--------------------------GACACA .................((((..(((.((((((((((((...))).))))))))))))..))))..((((...))--------------------------)).... ( -23.80) >consensus AAAUUAAGUAAAUAAAAUUGUUGCCGCUGUCAGGCGCAUAAAGUGACGCCUGACACGGACACGGAC_CGAACACG___________________GACAUG_GACACA .....................(((((.(((((((.((((...))).).)))))))))).)).............................................. (-16.24 = -16.18 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:42 2006