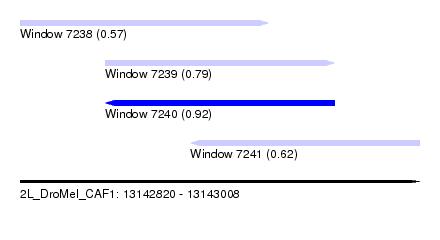
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,142,820 – 13,143,008 |
| Length | 188 |
| Max. P | 0.915613 |

| Location | 13,142,820 – 13,142,937 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -23.08 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13142820 117 + 22407834 AUAGAUUAAUCCAAACAGCACCAGCAGUCGGAGAAUGGAUUGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUGGGCUUUUAUAGCCUU--- ................(((.(((((((((((((....(((((.(........))))))..))))))))..(((((((((.....))))))))).....))))))))...........--- ( -37.10) >DroSec_CAF1 9891 117 + 1 AUAGAUUAAUCCAAACAGCACCAGCAGUCGGAGAAUGGAUUGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU--- .((((((((((......((....))....((((((((.....))))))))....)))))).))))(((..(((((((((.....)))))))))....)))..((((.....))))..--- ( -34.20) >DroSim_CAF1 10695 117 + 1 AUAGAUUAAUCCAAACAGCACCAGCAGUCGGAGAAUGGAUUGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU--- .((((((((((......((....))....((((((((.....))))))))....)))))).))))(((..(((((((((.....)))))))))....)))..((((.....))))..--- ( -34.20) >DroYak_CAF1 10053 117 + 1 AUAGAUCAAUUCAAACAGCACCAGCAGUCGGAGAAUGGAUUGCAUUUUUCUUUCGAUUAGCUCUGGCUGCAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUA--- ................(((....((((((((((....(((((.(........))))))..))))))))))(((((((((.....))))))))).....))).((((.....))))..--- ( -38.80) >DroAna_CAF1 9400 118 + 1 ACAGAUUAAUCCCAACAGCACCAAGAGACUGAUAUUUGCUUGCAUUU-UGUUUCUUUCAGCUUUCCGUGUAAUUGACUGAAGGU-GCUCGAUUGCAGAACCAGGCUUUUAUAGACACACU .(((.(((((..((..(((...(((((((.((....((....)).))-.)))))))...))).....))..))))))))...((-(((.((..((........))..))..)).)))... ( -18.70) >consensus AUAGAUUAAUCCAAACAGCACCAGCAGUCGGAGAAUGGAUUGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU___ .......................((((((........))))))...............(((((.((....(((((((((.....))))))))))).))))).((((.....))))..... (-23.08 = -23.76 + 0.68)

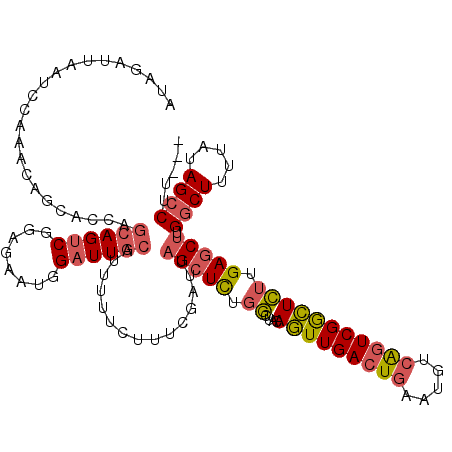
| Location | 13,142,860 – 13,142,968 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13142860 108 + 22407834 UGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUGGGCUUUUAUAGCCUU------GCCCUGGGUUAUCAG------CAGUUCUAAGUGGUUG ........................(((((((((((((((.....)))))))))...((((((((((.....))))..------...((((....))))------))))))...)))))). ( -32.40) >DroSec_CAF1 9931 111 + 1 UGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU------GCCCUGGGUUAUCAGCA---GCAGUUCUAAGUGGUUG .........(((..((((.(((((((..(((((((((((.....)))))))))((.(.((..((((.....))))..------)).).))))..)))).)---))))))..)))...... ( -32.40) >DroSim_CAF1 10735 111 + 1 UGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU------GCCCUGGGUUAUCAGCA---GCAGUUCUAAGUGGUUG .........(((..((((.(((((((..(((((((((((.....)))))))))((.(.((..((((.....))))..------)).).))))..)))).)---))))))..)))...... ( -32.40) >DroEre_CAF1 11850 104 + 1 ----UUUUUCUUUCGAUUAGCUCAGGCUACAGUUGACUGAAUGUCAGUUGGUUCUUUAGCUUGGCUUUUAUAGCCUA------GCUCUGGGCUAUCAG------CAGUGUUAAAUGGCUG ----.............((((((((....((..((((.....))))..)).......((((.((((.....)))).)------))))))))))).(((------(.((.....)).)))) ( -32.90) >DroYak_CAF1 10093 108 + 1 UGCAUUUUUCUUUCGAUUAGCUCUGGCUGCAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUA------GCCCUGGGUUAUCAG------CAGUUCUAAAUGAUUG .............((((((.....(((((((((((((((.....)))))))))((.(.(((.((((.....)))).)------)).).)).......)------))))).....)))))) ( -33.50) >DroAna_CAF1 9440 118 + 1 UGCAUUU-UGUUUCUUUCAGCUUUCCGUGUAAUUGACUGAAGGU-GCUCGAUUGCAGAACCAGGCUUUUAUAGACACACUCUGGGUCUGAGUUAUCAGCAAGUGCCAUUCUAUAUGUCUA ((((..(-((...((((((((.............).))))))).-...))).)))).....((((...((((((..((((....(.((((....))))).))))....)))))).)))). ( -25.92) >consensus UGCAUUUUUCUUUCGAUUAGCUCUGGCUACAGUUGACUGAAUGUCAGUCGGCUCUUGAGCUUGGCUUUUAUAGCCUU______GCCCUGGGUUAUCAG______CAGUUCUAAAUGGUUG ..............(((.(((((.(((...(((((((((.....))))))))).........((((.....))))........)))..))))))))........................ (-21.51 = -21.10 + -0.41)

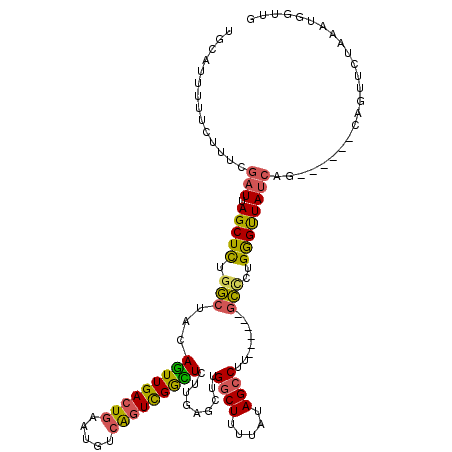
| Location | 13,142,860 – 13,142,968 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -14.58 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13142860 108 - 22407834 CAACCACUUAGAACUG------CUGAUAACCCAGGGC------AAGGCUAUAAAAGCCCAGCUCAAGAGCCGACUGACAUUCAGUCAACUGUAGCCAGAGCUAAUCGAAAGAAAAAUGCA .......((((..(((------(((........((((------..((((.....))))..)))).......(((((.....))))).....))).)))..))))((....))........ ( -28.50) >DroSec_CAF1 9931 111 - 1 CAACCACUUAGAACUGC---UGCUGAUAACCCAGGGC------AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAUUCAGUCAACUGUAGCCAGAGCUAAUCGAAAGAAAAAUGCA .......((((..((((---(((.(........((((------..((((.....))))..)))).......(((((.....)))))..).)))).)))..))))((....))........ ( -31.60) >DroSim_CAF1 10735 111 - 1 CAACCACUUAGAACUGC---UGCUGAUAACCCAGGGC------AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAUUCAGUCAACUGUAGCCAGAGCUAAUCGAAAGAAAAAUGCA .......((((..((((---(((.(........((((------..((((.....))))..)))).......(((((.....)))))..).)))).)))..))))((....))........ ( -31.60) >DroEre_CAF1 11850 104 - 1 CAGCCAUUUAACACUG------CUGAUAGCCCAGAGC------UAGGCUAUAAAAGCCAAGCUAAAGAACCAACUGACAUUCAGUCAACUGUAGCCUGAGCUAAUCGAAAGAAAAA---- ((((...........)------))).((((.(((.((------((((((.....))))............((..((((.....))))..))))))))).)))).((....))....---- ( -27.70) >DroYak_CAF1 10093 108 - 1 CAAUCAUUUAGAACUG------CUGAUAACCCAGGGC------UAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAUUCAGUCAACUGCAGCCAGAGCUAAUCGAAAGAAAAAUGCA .......((((..(((------(((........((((------(.((((.....)))).))))).......(((((.....))))).....))).)))..))))((....))........ ( -32.70) >DroAna_CAF1 9440 118 - 1 UAGACAUAUAGAAUGGCACUUGCUGAUAACUCAGACCCAGAGUGUGUCUAUAAAAGCCUGGUUCUGCAAUCGAGC-ACCUUCAGUCAAUUACACGGAAAGCUGAAAGAAACA-AAAUGCA ......((((((...((((((.((((....)))).....)))))).))))))...((.((.((((((......))-...((((((..............)))))))))).))-....)). ( -23.34) >consensus CAACCACUUAGAACUG______CUGAUAACCCAGGGC______AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAUUCAGUCAACUGUAGCCAGAGCUAAUCGAAAGAAAAAUGCA .......((((...........(((......)))...........(((((((........(((....))).(((((.....)))))...)))))))....))))((....))........ (-14.58 = -15.83 + 1.25)


| Location | 13,142,900 – 13,143,008 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.14 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -9.04 |
| Energy contribution | -10.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13142900 108 - 22407834 CACUCACCUCUGUCUAAAAGUAUACAUAUCACCUUUAAACCAACCACUUAGAACUG------CUGAUAACCCAGGGC------AAGGCUAUAAAAGCCCAGCUCAAGAGCCGACUGACAU ((.((..((((...............(((((.(......................)------.))))).....((((------..((((.....))))..)))).))))..)).)).... ( -17.35) >DroSec_CAF1 9971 111 - 1 CACUCACCUCAGUCUAAAAGUGUACAUAUCACCUUUAAACCAACCACUUAGAACUGC---UGCUGAUAACCCAGGGC------AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAU ........((((((((((.(((.......))).)))).........(((.((.((((---(.(((......))))))------..((((.....)))).)).)))))....))))))... ( -23.50) >DroSim_CAF1 10775 100 - 1 CACUCACCUCAGUCUAAAAGUGUA-----------UAAACCAACCACUUAGAACUGC---UGCUGAUAACCCAGGGC------AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAU ........((((((...(((((..-----------.........))))).....(((---(.(((......))))))------).((((.....))))..(((....))).))))))... ( -23.30) >DroEre_CAF1 11886 108 - 1 UACUCACCUCAAUCUAAGAGUAUGCAUAUAACCUUUAAACCAGCCAUUUAACACUG------CUGAUAGCCCAGAGC------UAGGCUAUAAAAGCCAAGCUAAAGAACCAACUGACAU (((((............)))))..........(((((...((((...........)------)))..........((------(.((((.....)))).))))))))............. ( -18.00) >DroYak_CAF1 10133 96 - 1 CACUCACCUCAGUCUAAAAGU------------CUUAAAGCAAUCAUUUAGAACUG------CUGAUAACCCAGGGC------UAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAU ........((((((.....(.------------.(((.((((.((.....))..))------))..)))..).((((------(.((((.....)))).))))).......))))))... ( -25.00) >DroAna_CAF1 9479 113 - 1 CACUCACAUUAAUU----GGUAUAC-UACUACCUAUCUA-UAGACAUAUAGAAUGGCACUUGCUGAUAACUCAGACCCAGAGUGUGUCUAUAAAAGCCUGGUUCUGCAAUCGAGC-ACCU ..............----((((...-...))))....((-(((((((((....(((......((((....))))..)))..)))))))))))...((.(((((....))))).))-.... ( -23.70) >consensus CACUCACCUCAGUCUAAAAGUAUAC_UAU_ACCUUUAAACCAACCACUUAGAACUG______CUGAUAACCCAGGGC______AAGGCUAUAAAAGCCAAGCUCAAGAGCCGACUGACAU ........((((((................................................(((......)))...........((((.....)))).............))))))... ( -9.04 = -10.10 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:54 2006