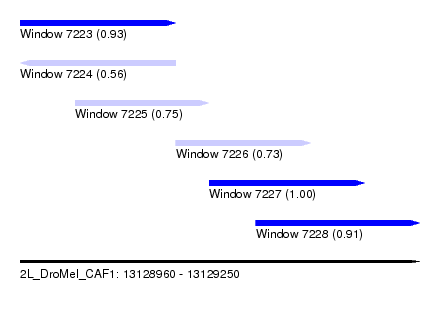
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,128,960 – 13,129,250 |
| Length | 290 |
| Max. P | 0.995831 |

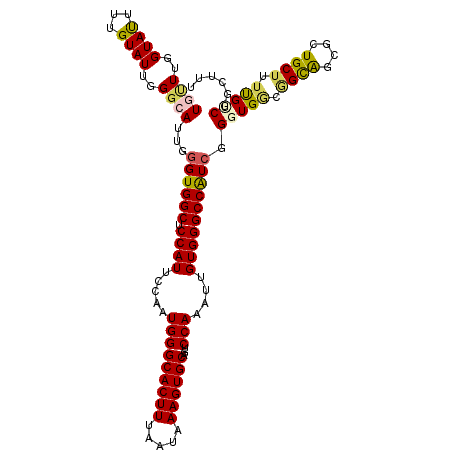
| Location | 13,128,960 – 13,129,073 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -30.90 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13128960 113 + 22407834 UGUUUGGUAUUUUGUAUUGGGCAUUGGGUGGCUCCAUUCCAAUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGGCGGCAGCGCUGCUCUUGCCCGCUUU ..................(((((...((((((.((((.....(((((((((.....))))))...)))....))))))))))(((..(((....)))..))).)))))..... ( -44.30) >DroSec_CAF1 21893 113 + 1 UGCUUGGUAUUUUGUAUUGGGCAUUGGGUGGCUCCAUUCCAAUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGUUGCUAGCAGCGCUGCUUUCGUCGGCUUU ((((..((((...))))..))))...((((((.((((.....(((((((((.....))))))...)))....)))))))))).((((....))))(((((....).))))... ( -39.70) >DroSim_CAF1 22981 113 + 1 UGCUUGGUAUUUUGUAUUGGGCAUUGGGUGGCUCCAUUCCAAUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCCGUUGGUGGCAGCGCUGCUUUCGUCGGCUUU ((((..((((...))))..))))..(((((((.((((.....(((((((((.....))))))...)))....)))))))))))((((..((.(((...))).))..))))... ( -43.40) >DroEre_CAF1 23676 113 + 1 UAUUGGGUAUUGGGUAUCGGCCAUUGGGUGGCUCCAUUCCGUUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCGUCGGGUGGCGGCAGCCCUGCUGUUGUCGGCUUU ...........((((..(.((((((.(..(((.((((....((((((((((.....))))))...))))...)))))))..).)))))).)..)))).((((....))))... ( -46.50) >DroYak_CAF1 22696 113 + 1 UAUUUCGUACUUGGUAUUGGGCAUUGAGUGGCUCCAUUCCAAUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGACGGUGGCCCUGCAUUUGCCGGCUUU ..........((((((((((((...(((((....)))))......((((((.....)))))).))))))..((((((((((((.....))))))))).)))..)))))).... ( -44.40) >consensus UGUUUGGUAUUUUGUAUUGGGCAUUGGGUGGCUCCAUUCCAAUGGGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGGCGGCAGCGCUGCUUUUGUCGGCUUU ((((..((((...))))..))))...((((((.((((.....(((((((((.....))))))...)))....)))))))))).(((((.((((....)))).)))))...... (-30.90 = -31.14 + 0.24)

| Location | 13,128,960 – 13,129,073 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13128960 113 - 22407834 AAAGCGGGCAAGAGCAGCGCUGCCGCCACCCGAUGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAUUGGAAUGGAGCCACCCAAUGCCCAAUACAAAAUACCAAACA .....(((((.(.(((....)))).(((.(((((((.(((.....)))...((((((.....)))))))))))))..)))..........))))).................. ( -32.40) >DroSec_CAF1 21893 113 - 1 AAAGCCGACGAAAGCAGCGCUGCUAGCAACCGAUGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAUUGGAAUGGAGCCACCCAAUGCCCAAUACAAAAUACCAAGCA ..((((..(....)..).)))(((.....(((((((.(((.....)))...((((((.....)))))))))))))..(((.((........))))).............))). ( -30.50) >DroSim_CAF1 22981 113 - 1 AAAGCCGACGAAAGCAGCGCUGCCACCAACGGAUGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAUUGGAAUGGAGCCACCCAAUGCCCAAUACAAAAUACCAAGCA ...((.(.(....)).))(((((((((...)).)))).......((((...((((((.....)))))).((((((..((....)).)))))).))))............))). ( -29.20) >DroEre_CAF1 23676 113 - 1 AAAGCCGACAACAGCAGGGCUGCCGCCACCCGACGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAACGGAAUGGAGCCACCCAAUGGCCGAUACCCAAUACCCAAUA ...((........)).((((((.((.....)).)))))).....((((...((((((.....)))))).....((.((.(.((((.....))))).)).))......)))).. ( -31.00) >DroYak_CAF1 22696 113 - 1 AAAGCCGGCAAAUGCAGGGCCACCGUCACCCGAUGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAUUGGAAUGGAGCCACUCAAUGCCCAAUACCAAGUACGAAAUA .....((((....)).((((((.((.....)).))))))...((((((...((((((.....))))))..(((((.((.(((...))).))..))))).)))))).))..... ( -33.20) >consensus AAAGCCGACAAAAGCAGCGCUGCCGCCACCCGAUGGCCCACAAUUUGGACUGCACUUUAUUAAAGUGCCCAUUGGAAUGGAGCCACCCAAUGCCCAAUACAAAAUACCAAACA ...((........)).(.(((.(((....(((((((.(((.....)))...((((((.....)))))))))))))..)))))))............................. (-20.86 = -21.10 + 0.24)

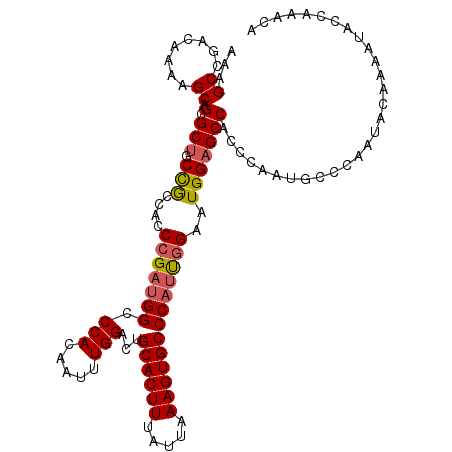
| Location | 13,129,000 – 13,129,097 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.25 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13129000 97 + 22407834 AAUG-------GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGGCGGCAGCGCUGCUCUUGCCCGCUUUGUUGUUGCUGU---------CGGUGGU--GCUGAU----- ..((-------(((((((.....))))))...)))......(.(((((((.(..(((((((((..((........))..)))))))))..)---------)))))))--.)....----- ( -37.10) >DroSec_CAF1 21933 97 + 1 AAUG-------GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGUUGCUAGCAGCGCUGCUUUCGUCGGCUUUGUUGUUGCUGG---------CGAUGGU--GCUGAU----- ....-------.((((((.....)))))).(((((.....)))))(((((.(((((((((((((.(((......)))...).)))))))))---------)))))))--).....----- ( -35.80) >DroSim_CAF1 23021 97 + 1 AAUG-------GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCCGUUGGUGGCAGCGCUGCUUUCGUCGGCUUUGUUGUUGCUGU---------CGGUGGU--GCUGAU----- ((((-------(((((((.....)))))).(((((.....)))))...))))).....((((((..(...((.((((.........)))).---------)))..))--))))..----- ( -35.80) >DroEre_CAF1 23716 97 + 1 GUUG-------GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCGUCGGGUGGCGGCAGCCCUGCUGUUGUCGGCUUUGUUGUUGCUGU---------CGGUGGU--GCUGAU----- .(((-------(((((((.....))))))...)))).....(.((((.(.((..((((((((...((((....))))...))))))))..)---------).)))))--.)....----- ( -37.60) >DroYak_CAF1 22736 102 + 1 AAUG-------GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGACGGUGGCCCUGCAUUUGCCGGCUUUGUUGUUGCUGU---------CAGUGGU--GCUGAUGCGGC ....-------.((((((.....))))))(((((((((.(((((((((((.....))))))))).)))))))..)))).....(((((.((---------(((....--.)))))))))) ( -41.20) >DroAna_CAF1 28979 107 + 1 GAUGAGCCGAGGGUACUUUAAUAAAGUGCAGUCCAAAUUGAGGUCCACCAG------GCAGCCGGGCUUA--UGGUUUUCGUUGUUGCUGUUAUUUCAAGCGGUGGUAGGCAGAC----- .....(((((..((((((.....))))))..))...........(((((.(------(((((.((((...--..))))..))))))(((.........))))))))..)))....----- ( -29.40) >consensus AAUG_______GGCACUUUAAUAAAGUGCAGUCCAAAUUGUGGGCCAUCGGGUGGCGGCAGCCCUGCUUUUGUCGGCUUUGUUGUUGCUGU_________CGGUGGU__GCUGAU_____ ............((((((.....))))))...(((.....)))(((((((....((((((((...((........))......)))))))).........)))))))............. (-20.30 = -20.25 + -0.05)

| Location | 13,129,073 – 13,129,171 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.91 |
| Mean single sequence MFE | -47.63 |
| Consensus MFE | -32.93 |
| Energy contribution | -33.82 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13129073 98 + 22407834 GUUGUUGCUGU---------CGGUGGU--GCUGAU-----------GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAU ....(((((..---------.((((((--(.((((-----------((.((((((((((....)))).)))))).))))))..)))))))(((.(((.....))).))).)))))..... ( -44.80) >DroSec_CAF1 22006 98 + 1 GUUGUUGCUGG---------CGAUGGU--GCUGAU-----------GCGGCGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAU ....(((((((---------(((((((--..((((-----------((.((((((((((....)))).)))))).)))))))))))))))(((.(((.....))).))).)))))..... ( -53.60) >DroSim_CAF1 23094 98 + 1 GUUGUUGCUGU---------CGGUGGU--GCUGAU-----------GCGGCGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAU ....(((((..---------.((((((--(.((((-----------((.((((((((((....)))).)))))).))))))..)))))))(((.(((.....))).))).)))))..... ( -49.60) >DroEre_CAF1 23789 98 + 1 GUUGUUGCUGU---------CGGUGGU--GCUGAU-----------GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAA ....(((((..---------.((((((--(.((((-----------((.((((((((((....)))).)))))).))))))..)))))))(((.(((.....))).))).)))))..... ( -44.80) >DroYak_CAF1 22809 109 + 1 GUUGUUGCUGU---------CAGUGGU--GCUGAUGCGGCGGUGAUGCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAACAAU (((((((((((---------(..((((--((....(((((((((((((.((((((((((....)))).)))))).))))))....))))))).)))).))......)))).)).)))))) ( -50.30) >DroAna_CAF1 29051 109 + 1 GUUGUUGCUGUUAUUUCAAGCGGUGGUAGGCAGAC-----------CUCGGCCUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUAGGAAAAGUGGGGCAGGCAAAUAAU .((((((((.(..((((.(((((((((((...(((-----------(((((...(((((....)))))..))).)).)))..)))))))))))....))))...).)))).))))..... ( -42.70) >consensus GUUGUUGCUGU_________CGGUGGU__GCUGAU___________GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAU ....(((((............((((((....((((...........((.((((((((((....)))).)))))).))))))...))))))(((.(((.....))).))).)))))..... (-32.93 = -33.82 + 0.89)

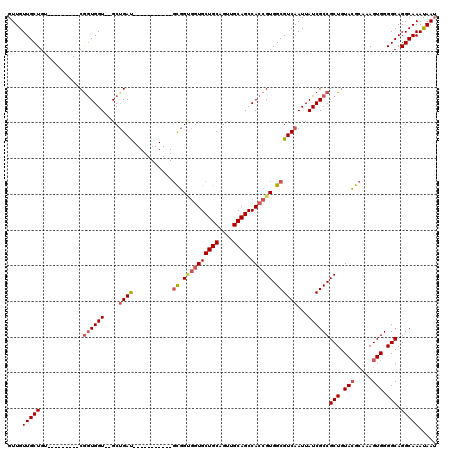
| Location | 13,129,097 – 13,129,210 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.45 |
| Mean single sequence MFE | -42.11 |
| Consensus MFE | -39.39 |
| Energy contribution | -39.34 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13129097 113 + 22407834 ------GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ------(((..((((((((....)))).))))((((((........))))))....)))..((((((((..((........-.....))))))....(.((((....)))))....)))) ( -41.22) >DroSec_CAF1 22030 113 + 1 ------GCGGCGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ------(((((((((((((....)))).))))))(((((..(((((.((((((.(((.....))).))).)))..))))).-.....)))))....)))((((....))))......... ( -42.80) >DroSim_CAF1 23118 113 + 1 ------GCGGCGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ------(((((((((((((....)))).))))))(((((..(((((.((((((.(((.....))).))).)))..))))).-.....)))))....)))((((....))))......... ( -42.80) >DroEre_CAF1 23813 114 + 1 ------GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAAAAAAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ------(((..((((((((....)))).))))((((((........))))))....)))..((((((((..((..............))))))....(.((((....)))))....)))) ( -41.14) >DroYak_CAF1 22838 119 + 1 GGUGAUGCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAACAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ..((((((.((((((((((....)))).)))))).))))))......((((((.(((.....))).))).)))........-.....((((......))((((....))))....))... ( -42.50) >DroAna_CAF1 29086 113 + 1 ------CUCGGCCUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUAGGAAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGCCGUCAGGCUCAAAGCACU ------....(((((((.((.((....((((.((((((........)))))).)).))..)).)).)))))))........-.....((((......))((((....))))....))... ( -42.20) >consensus ______GCGGUGGUGCUGCAGUUGCAGCCACCGUGGCGUCAAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA_AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACU ......(((..((((((((....)))).))))((((((........))))))....)))..((((((((..((..............))))))....(.((((....)))))....)))) (-39.39 = -39.34 + -0.05)

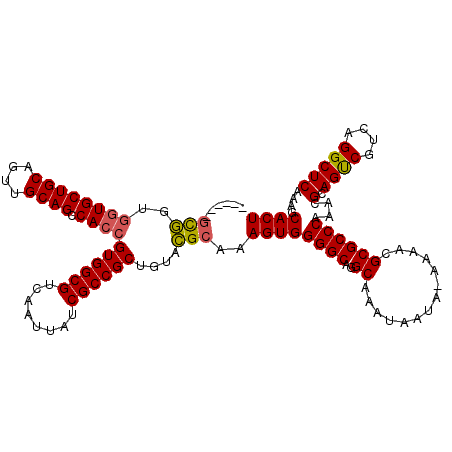
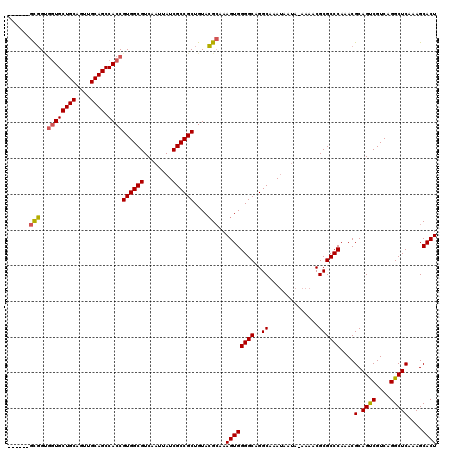
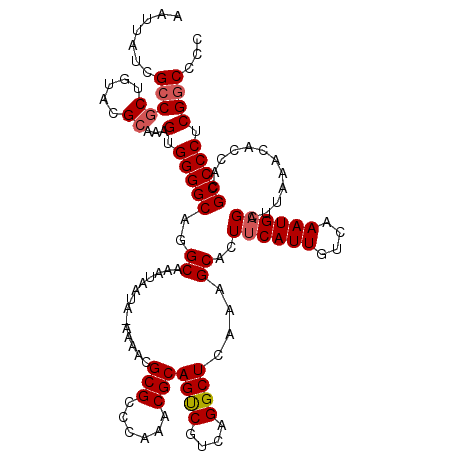
| Location | 13,129,131 – 13,129,250 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -28.37 |
| Energy contribution | -28.73 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13129131 119 + 22407834 AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAACACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((.((.........-.....(((......))).......(((((((....((......))....))))))).....))..))))).))))... ( -31.10) >DroSec_CAF1 22064 119 + 1 AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAAAACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((.((.........-.....(((......))).......(((((((....((......))....))))))).....))..))))).))))... ( -31.10) >DroSim_CAF1 23152 119 + 1 AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAACACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((.((.........-.....(((......))).......(((((((....((......))....))))))).....))..))))).))))... ( -31.10) >DroEre_CAF1 23847 120 + 1 AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAAAAAAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAGCACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((..................(((......)))....((...(((..((((...(((((....))))))))).)))...))))))).))))... ( -32.30) >DroYak_CAF1 22878 119 + 1 AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAACAAUA-AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAACACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((.((.........-.....(((......))).......(((((((....((......))....))))))).....))..))))).))))... ( -31.10) >DroAna_CAF1 29120 116 + 1 AAUUAUCGCCGCUGUAGGAAAAGUGGGGCAGGCAAAUAAUA-AAAACGCGCCCAAACGCAGCCGUCAGGCUCAAAGCACUCCAUUGUCAAAUGAGUGCAAAC---ACGCCCCUCGACCCC .....((.((......))....(.(((((............-.....(((......)))((((....))))....((((((.(((....)))))))))....---..))))).))).... ( -33.00) >consensus AAUUAUCGCCGCUGUACGCAAAGUGGGGCAGGCAAAUAAUA_AAAACGCGCCCAAACGCAGUCGUCAGGCUCAAAGCACUUCAUUGUCAAAUGAGCUUAAACACCACGCCCCUCGGCCCC .......(((((.....))...(.(((((..((..............(((......)))((((....))))....))..((((((....))))))............))))).))))... (-28.37 = -28.73 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:42 2006