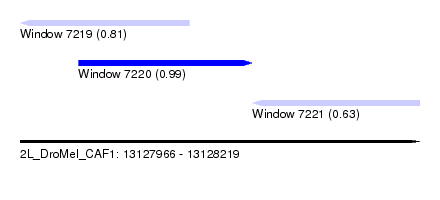
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,127,966 – 13,128,219 |
| Length | 253 |
| Max. P | 0.993694 |

| Location | 13,127,966 – 13,128,073 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -27.21 |
| Consensus MFE | -23.05 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
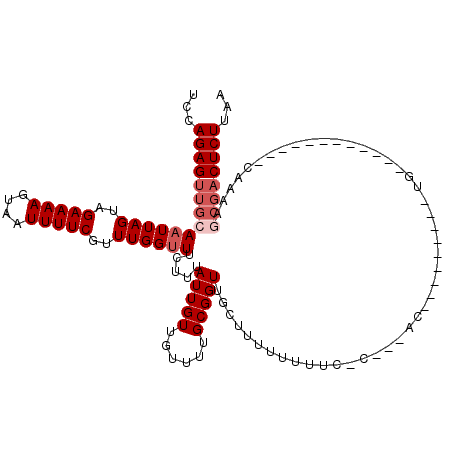
>2L_DroMel_CAF1 13127966 107 - 22407834 CCCUUUGUUCGACUGGUUAGUUGAA-AACGCAAAUACAUUCGUACAUCCGAAACUAACCGAAACUUGGUUGGAUGCAUGAUUAAUGUCCGCAUUGAGUCAUGUCCGAG ...(((((((((((....)))))).-...)))))....((((......)))).((((((((...))))))))..((((((((((((....)))))).))))))..... ( -25.80) >DroSec_CAF1 20934 87 - 1 CCCUUUGUUCGACUGGUUAGUUGAA-AACACA--------------------ACUAACCGAAACUUGGUUGGAUGCAUGAUUAAUGUCCGCAUUGAGUCAUGUCCGAG ......(((((((((((((((((..-....))--------------------))))))).......))))))))((((((((((((....)))))).))))))..... ( -27.51) >DroSim_CAF1 22018 87 - 1 CCCUUUGUUCGACUGGUUAGUUGAA-AACACA--------------------ACUAACCGAAACUUGGUUGGAUGCAUGAUUAAUGUCCGCAUUGAGUCAUGUCCGAG ......(((((((((((((((((..-....))--------------------))))))).......))))))))((((((((((((....)))))).))))))..... ( -27.51) >DroYak_CAF1 21367 102 - 1 CCCUUUGUUUGGCUGGUUAGUUGGAAAACGCACAUACAUCUGUACG------ACUAACCGAAACUCGGUUGGAUGCAUGAUUAAUGUCCGCAUUGAGUCAUGUCCGAG .((.......)).((((((((((.....................))------))))))))...(((((......((((((((((((....)))))).))))))))))) ( -28.00) >consensus CCCUUUGUUCGACUGGUUAGUUGAA_AACACA____________________ACUAACCGAAACUUGGUUGGAUGCAUGAUUAAUGUCCGCAUUGAGUCAUGUCCGAG .......(((((((....)))))))............................((((((((...))))))))..((((((((((((....)))))).))))))..... (-23.05 = -22.30 + -0.75)


| Location | 13,128,003 – 13,128,113 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -19.65 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13128003 110 + 22407834 --------CAACCAAGUUUCGGUUAGUUUCGGAUGUACGAAUGUAUUUGCGU-U-UUCAACUAACCAGUCGAACAAAGGGUGUUAGAAUGCUUUUUGUUUCAGCAUCGCUGAGCAGAGAU --------.......(((((((((((((..(((((((..........)))))-)-)..)))))))).((..((((((((((((....))))))))))))(((((...))))))).))))) ( -29.70) >DroSec_CAF1 20971 90 + 1 --------CAACCAAGUUUCGGUUAGU--------------------UGUGU-U-UUCAACUAACCAGUCGAACAAAGGGUGUUAGAAUGCUUUUUGUUUCAGCAUCGCUGAGCAGAGAU --------.......((((((((((((--------------------((...-.-..))))))))).((..((((((((((((....))))))))))))(((((...))))))).))))) ( -28.90) >DroSim_CAF1 22055 90 + 1 --------CAACCAAGUUUCGGUUAGU--------------------UGUGU-U-UUCAACUAACCAGUCGAACAAAGGGUGUUAGAAUGCUUUUUGUUUCAGCAUCGCUGAGCAGAGAU --------.......((((((((((((--------------------((...-.-..))))))))).((..((((((((((((....))))))))))))(((((...))))))).))))) ( -28.90) >DroYak_CAF1 21404 105 + 1 --------CAACCGAGUUUCGGUUAGU------CGUACAGAUGUAUGUGCGU-UUUCCAACUAACCAGCCAAACAAAGGGUGUUAGAAUGCUUUUUGUUUCAGCAUCGCUGAGCAGAGAU --------.......((((((((((((------((((((......)))))).-......))))))).((..((((((((((((....))))))))))))(((((...))))))).))))) ( -32.51) >DroAna_CAF1 28092 101 + 1 AGACCACACAACAAAGUCUUGAAUAUA------------------UAUGGAUAU-UUGAAAAAGCUAAACGAACAAAGAGUGUUAGAAUACUUUUUGUUUCAGCAUCGUUGAGCAGAGAU ...............(((((((((((.------------------.....))))-))......(((.((((((((((((((((....))))))))))).......))))).))).))))) ( -21.61) >consensus ________CAACCAAGUUUCGGUUAGU__________________U_UGCGU_U_UUCAACUAACCAGUCGAACAAAGGGUGUUAGAAUGCUUUUUGUUUCAGCAUCGCUGAGCAGAGAU ...............((((((((((((................................))))))).....((((((((((((....))))))))))))(((((...)))))...))))) (-19.65 = -19.17 + -0.48)

| Location | 13,128,113 – 13,128,219 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -13.57 |
| Energy contribution | -14.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13128113 106 - 22407834 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUGCUUUUUUU-C-C---ACCGACUCCCCUG---------CCCCAAAAGCGACUCUUAA ...((((((((........(((((....))))).(((((..........((......))(((.(........-)-.---)))..........---------..))))).))))))))... ( -21.00) >DroSec_CAF1 21061 95 - 1 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUGCUUUUUUUUC-C---AC---------UG------------CAAAAGCGACUCUUAA ...((((((((((((((..(((((....)))))..))))))............(((((((((............-.---))---------))------------)))))))))))))... ( -24.42) >DroSim_CAF1 22145 95 - 1 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUGCUUUUUUUUC-C---AC---------UG------------CAAAAGCGACUCUUAA ...((((((((((((((..(((((....)))))..))))))............(((((((((............-.---))---------))------------)))))))))))))... ( -24.42) >DroEre_CAF1 22480 100 - 1 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUUGCUUUUUUCCGC---ACCC-----------------CCCCCAAAACCGACUCUUAA ...(((((((.((((((..(((((....)))))..))))))...........((((((.((..(((........))---)..)-----------------)...)))))))))))))... ( -21.20) >DroYak_CAF1 21509 119 - 1 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUGCUUUUUUUUCGCCUCACCCA-UCCCCUGACAGCCUCCACCAAAAAGCGACUCUUAA ...((((((((........(((((....))))).((((((.........((((((.((.(((.((.........))...)))))-......))))))....))))))..))))))))... ( -26.12) >DroAna_CAF1 28193 87 - 1 UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGCUUCUUUAUUGUUGUUUUGCGGUUGCUGGUUAUUU-C---GU---------UC--------------------UCUCUCAA .((((((..((((.(((((((.((((............)))).))))))).....))))..)).))))......-.---..---------..--------------------........ ( -15.30) >consensus UCCAGAGUUGCAAUUAGUAGAAAAGUAAUUUUCGUUUGGUUUCUUUAUUGUUGUUUUGCGGUUGCUUUUUUUUC_C___AC_________UG____________CAAAAGCGACUCUUAA ...((((((((((((((..(((((....)))))..)))))).....(((((......)))))...............................................))))))))... (-13.57 = -14.57 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:35 2006