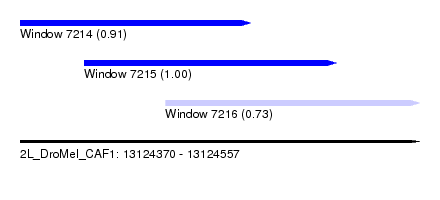
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,124,370 – 13,124,557 |
| Length | 187 |
| Max. P | 0.997501 |

| Location | 13,124,370 – 13,124,478 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
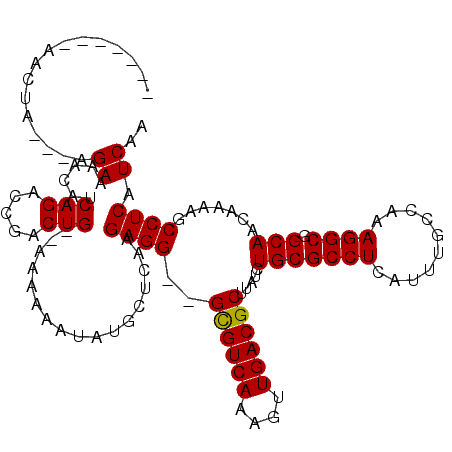
>2L_DroMel_CAF1 13124370 108 + 22407834 -------AACUA---AGAAAACAUCAGACCGACUGCAAAAAAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA -------.....---.((................(((........)))....((((--((((((....)))))).....(((((((..........)))).))).......)))).)).. ( -23.70) >DroSec_CAF1 17358 108 + 1 -------AACUA---AGAAAACAUCAGACCGACUG--AAAAAAUAUGCUCAAGAGGGGGUGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA -------.....---.((.....((((.....)))--)..............((((((((((((....))))))))...(((((((..........)))).))).......)))).)).. ( -25.10) >DroSim_CAF1 18456 106 + 1 -------AACUA---AGAAAACAUCAGACCGACUG--AAAAAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA -------.....---.((.....((((.....)))--)..............((((--((((((....)))))).....(((((((..........)))).))).......)))).)).. ( -25.00) >DroEre_CAF1 18987 109 + 1 -------AACUAAGAAGAAAGCCUCAGGCCGACUG--AAAAAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA -------......((.((..((((..(((.((.((--(........((.((.((((--((((((....)))))))).)))).)).))))).)))..))))(((.......))))).)).. ( -29.70) >DroYak_CAF1 17813 113 + 1 CAUAGGAAACUA---AGAAAGCAUCAGGCCGACUG--AAAAAAUAUGCUUAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA ..(((....)))---.(((((((((((.....)))--).......)))))..((((--((((((....)))))).....(((((((..........)))).))).......)))).)).. ( -32.91) >consensus _______AACUA___AGAAAACAUCAGACCGACUG__AAAAAAUAUGCUCAAGAGG__GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAA ................((......(((.....))).................((((..((((((....)))))).....(((((((..........)))).))).......)))).)).. (-21.44 = -21.28 + -0.16)

| Location | 13,124,400 – 13,124,518 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.13 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
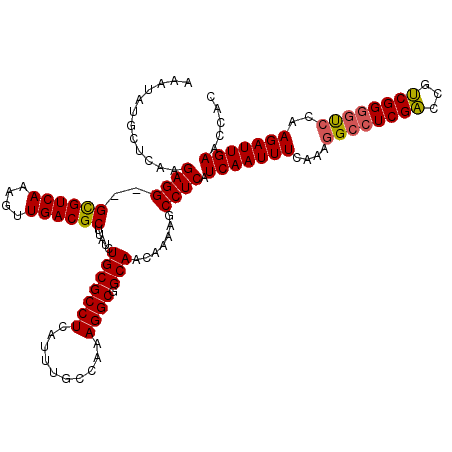
>2L_DroMel_CAF1 13124400 118 + 22407834 AAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCUAAGAUUGAACCAC ............((((--((((((....)))))).....(((((((..........)))).))).......)))).(((((((...((((((((.....)))))))).)))))))..... ( -38.80) >DroSec_CAF1 17386 120 + 1 AAAUAUGCUCAAGAGGGGGUGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCCAAGAUUGAACCAC ............((((((((((((....))))))))...(((((((..........)))).))).......)))).(((((((....(((((((.....)))))))..)))))))..... ( -37.20) >DroSim_CAF1 18484 118 + 1 AAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGUUCCAAGAUUGAACCAC ............((((--((((((....)))))).....(((((((..........)))).))).......)))).(((((((....((((.((.....))))))...)))))))..... ( -31.60) >DroEre_CAF1 19018 118 + 1 AAAUAUGCUCAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGGCCGCCGGGGCCCAAGAUUGAACCAC ............((((--((((((....)))))).....(((((((..........)))).))).......)))).(((((((....((((((((...))))))))..)))))))..... ( -40.90) >DroYak_CAF1 17848 118 + 1 AAAUAUGCUUAAGAGG--GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGAGCACAAGAUUGAACCAC ............((((--((((((....)))))).....(((((((..........)))).))).......)))).(((((((.....((..((.....))..))...)))))))..... ( -31.90) >consensus AAAUAUGCUCAAGAGG__GCGUCAAAGUUGACGCUUAUCUGCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCCAAGAUUGAACCAC ............((((..((((((....)))))).....(((((((..........)))).))).......)))).(((((((....((((((((...))))))))..)))))))..... (-33.78 = -33.66 + -0.12)

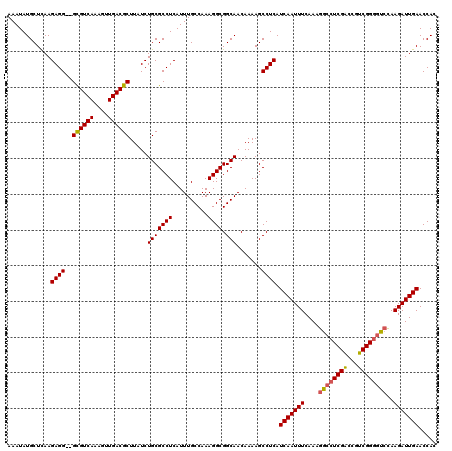
| Location | 13,124,438 – 13,124,557 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -33.94 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13124438 119 + 22407834 GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCUAAGAUUGAACCACUGACAGCUGUCUGCAGGAGGAGCAAAUACGA-UGGAUACU ((.((((...(((....((((((.......)))...(((((((...((((((((.....)))))))).)))))))..........)))....))).)))).))........-........ ( -37.70) >DroSec_CAF1 17426 119 + 1 GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCCAAGAUUGAACCACUGACAGCUGUCUGCAGGAGGAGCAAAUGCGA-UGGAUACU ((.((((...(((...(((((((.......((((............)))).......((((((((.((....)).))).))))).)))))))))).)))).))........-........ ( -36.90) >DroSim_CAF1 18522 119 + 1 GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGUUCCAAGAUUGAACCACUGACAGCUGUCUGCAGGAGGAGCAAAUGCGA-UGGAUACU ((.((((...(((...(((((((.......((((............)))).......(((.(((((.......)))))...))).)))))))))).)))).))........-........ ( -34.80) >DroEre_CAF1 19056 117 + 1 GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGGCCGCCGGGGCCCAAGAUUGAACCACUGACAGCUGUCCGCAGGAGGAGCAGAUGGAACUAGAU--- ((.((((...(((....((((((.......)))...(((((((....((((((((...))))))))..)))))))..........)))....))).)))).))..............--- ( -39.80) >DroYak_CAF1 17886 119 + 1 GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGAGCACAAGAUUGAACCACUGACAGCUGUCUGCAGGAGGAGCAGAUAGCA-UGGAUGCU ((((((((...(((...)))(((.......)))...(((((((.....((..((.....))..))...))))))).....)))..(((((((((.......))))))))).-.)).))). ( -36.20) >consensus GCGCCUCAUUUGCCAAAGGCGGCAACAAAAGCCUCAUCAAUUUCAAAGGCCUCGACCGUCGGGGUCCAAGAUUGAACCACUGACAGCUGUCUGCAGGAGGAGCAAAUGCGA_UGGAUACU ((.((((...(((....((((((.......)))...(((((((....((((((((...))))))))..)))))))..........)))....))).)))).))................. (-33.94 = -33.98 + 0.04)

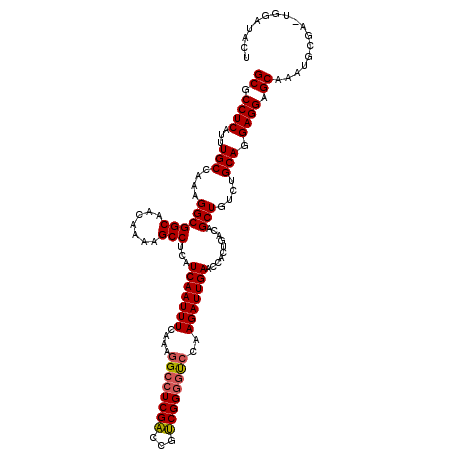
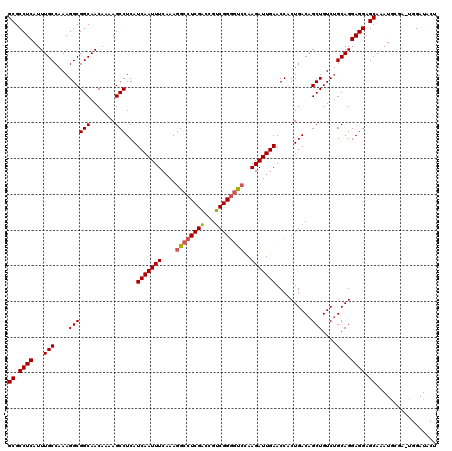
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:31 2006