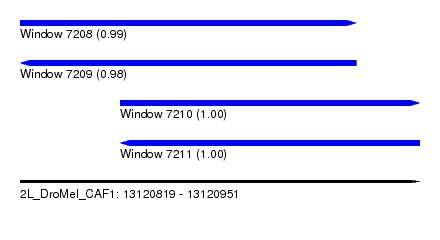
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,120,819 – 13,120,951 |
| Length | 132 |
| Max. P | 0.999912 |

| Location | 13,120,819 – 13,120,930 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
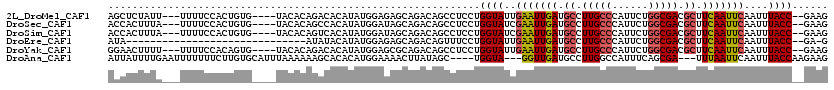
>2L_DroMel_CAF1 13120819 111 + 22407834 AGCUCUAUU---UUUUCCACUGUG----UACACAGACACAUAUGGAGAGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC--GAAG .(((.....---(((((((.((((----(......)))))..))))))).....)))....((((((((((((((.((.(((((......))))).)).)))))))))..))))--)... ( -36.30) >DroSec_CAF1 13737 111 + 1 ACCACUUUA---UUUUCCACUGUG----UACACAGCCACAUAUGGAUAGCAGACAGCCUCCUGGUAUCGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC--GAAG ....((((.---...((((.((((----(........)))))))))................((((..(((((((.((.(((((......))))).)).)))))))....))))--)))) ( -28.80) >DroSim_CAF1 14966 111 + 1 ACCACUUUA---UUUUCCACUGUG----UACACAGUCACAUAUGGAUAGCAGACAGCCUCCUGGUAUCGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC--GAAG ....((((.---......(((((.----...)))))........((((.(((........))).))))(((((((.((.(((((......))))).)).)))))))........--)))) ( -29.80) >DroEre_CAF1 15569 87 + 1 AUA------------------------------AUAUACAUAUGGAGAGCAGACAGUUUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC--GA-G ...------------------------------..........(((((........)))))((((((((((((((.((.(((((......))))).)).)))))))))..))))--).-. ( -27.00) >DroYak_CAF1 14117 111 + 1 GGAACUUUU---UUUUCCACAGUG----UACACAGACACAUAUGGAGCGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC--GAAG ((((.....---..))))...(((----(......))))....((((.((.....))))))((((((((((((((.((.(((((......))))).)).)))))))))..))))--)... ( -37.60) >DroAna_CAF1 21911 110 + 1 AUUAUUUUGAAUUUUUUUCUUGUGCAUUUAAAAAAGCACACAUGGAAAACUUAUAGC----UGGUA---GGUUGAUGCCUUGGCCAUUUCAGCGA---UUUAAUUCAAUUUACCAAGAAG ..((..(((((((.(((((((((((..........)))))...))))))......((----(((..---(((..(....)..)))...)))))..---...)))))))..))........ ( -24.50) >consensus ACCACUUUA___UUUUCCACUGUG____UACACAGACACAUAUGGAGAGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACC__GAAG ..............................................................((((..(((((((.((.(((((......))))).)).)))))))....))))...... (-16.51 = -16.93 + 0.42)


| Location | 13,120,819 – 13,120,930 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.88 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13120819 111 - 22407834 CUUC--GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCUCUCCAUAUGUGUCUGUGUA----CACAGUGGAAAA---AAUAGAGCU ((((--((((..(((((((((.((.(.(((......))).).)).))))))))))))).)))).......(((((((((.(((((......)----)))))))))...---....)))). ( -38.71) >DroSec_CAF1 13737 111 - 1 CUUC--GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCGAUACCAGGAGGCUGUCUGCUAUCCAUAUGUGGCUGUGUA----CACAGUGGAAAA---UAAAGUGGU ((((--((((..(((((((((.((.(.(((......))).).)).))))))))))))).))))((.....))..(((((.((((........----)))))))))...---......... ( -36.00) >DroSim_CAF1 14966 111 - 1 CUUC--GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCGAUACCAGGAGGCUGUCUGCUAUCCAUAUGUGACUGUGUA----CACAGUGGAAAA---UAAAGUGGU ((((--((((..(((((((((.((.(.(((......))).).)).))))))))))))).))))((.....))..(((((.((((........----)))))))))...---......... ( -34.70) >DroEre_CAF1 15569 87 - 1 C-UC--GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAAACUGUCUGCUCUCCAUAUGUAUAU------------------------------UAU .-..--((...((((((((((.((.(.(((......))).).)).))))))))))..(((....)))........))..........------------------------------... ( -25.60) >DroYak_CAF1 14117 111 - 1 CUUC--GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCGCUCCAUAUGUGUCUGUGUA----CACUGUGGAAAA---AAAAGUUCC ((((--((((..(((((((((.((.(.(((......))).).)).))))))))))))).))))((.....))..((((((.((((......)----)))))))))...---......... ( -37.00) >DroAna_CAF1 21911 110 - 1 CUUCUUGGUAAAUUGAAUUAAA---UCGCUGAAAUGGCCAAGGCAUCAACC---UACCA----GCUAUAAGUUUUCCAUGUGUGCUUUUUUAAAUGCACAAGAAAAAAAUUCAAAAUAAU ............(((((((...---..(((...(((((..(((......))---)....----))))).)))((((....(((((..........))))).))))..)))))))...... ( -18.40) >consensus CUUC__GGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCUCUCCAUAUGUGUCUGUGUA____CACAGUGGAAAA___AAAAGUGAU ......((((..(((((((((.((.(.(((......))).).)).))))))))))))).((((((.....))).)))........................................... (-18.88 = -20.88 + 2.00)

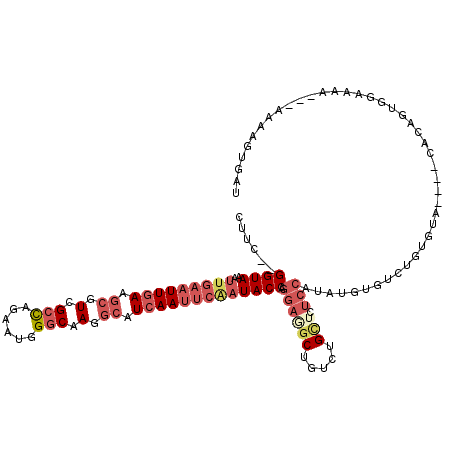
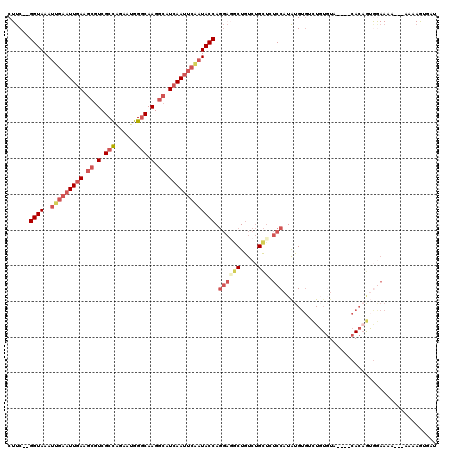
| Location | 13,120,852 – 13,120,951 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.30 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -33.46 |
| Energy contribution | -34.22 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13120852 99 + 22407834 UAUGGAGAGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGAAGGGGAUUCCUCUUCCAAAAAAA ...((((.((.....)))))).(((((((((((((.((.(((((......))))).)).)))))))))..))))(((((((...)))))))........ ( -39.00) >DroSec_CAF1 13770 96 + 1 UAUGGAUAGCAGACAGCCUCCUGGUAUCGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGAAGGGGAUUCCUCUUCCAAAA--- ....((((.(((........))).))))(((((((.((.(((((......))))).)).)))))))........(((((((...))))))).....--- ( -33.10) >DroSim_CAF1 14999 97 + 1 UAUGGAUAGCAGACAGCCUCCUGGUAUCGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGAAGGGGAUUCCUCUUCCAAAAA-- ....((((.(((........))).))))(((((((.((.(((((......))))).)).)))))))........(((((((...)))))))......-- ( -33.10) >DroEre_CAF1 15579 96 + 1 UAUGGAGAGCAGACAGUUUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGA-GGGGAUUCCUCUUCCGAAAA-- ..(((((((.....(((..((((((((((((((((.((.(((((......))))).)).)))))))))..)))).)-))..))).))).))))....-- ( -36.80) >DroYak_CAF1 14150 97 + 1 UAUGGAGCGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGAAGGGGAUUCCUCUUCCGAAAA-- ...((((.((.....)))))).(((((((((((((.((.(((((......))))).)).)))))))))..))))(((((((...)))))))......-- ( -39.00) >consensus UAUGGAGAGCAGACAGCCUCCUGGUAUUGAAUUGAUGCCUUGCCCAUUCUGGCGACGCUUCAAUUCAAUUUACCGAAGGGGAUUCCUCUUCCAAAAA__ ...((((.((.....)))))).((((.((((((((.((.(((((......))))).)).))))))))...))))(((((((...)))))))........ (-33.46 = -34.22 + 0.76)

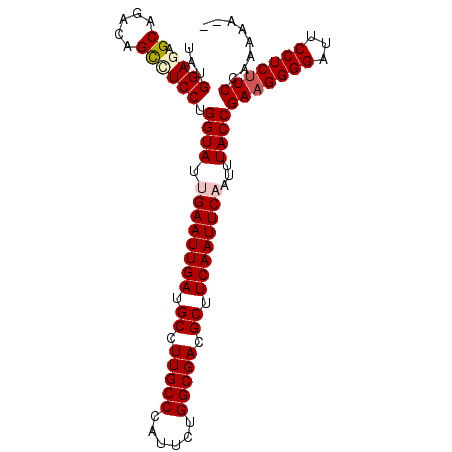
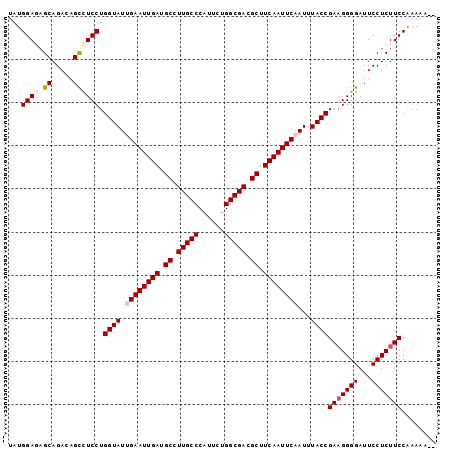
| Location | 13,120,852 – 13,120,951 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.30 |
| Mean single sequence MFE | -36.16 |
| Consensus MFE | -32.52 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.51 |
| SVM RNA-class probability | 0.999912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13120852 99 - 22407834 UUUUUUUGGAAGAGGAAUCCCCUUCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCUCUCCAUA ........((((.((....))))))((((..(((((((((.((.(.(((......))).).)).))))))))))))).((((((.....)).))))... ( -37.50) >DroSec_CAF1 13770 96 - 1 ---UUUUGGAAGAGGAAUCCCCUUCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCGAUACCAGGAGGCUGUCUGCUAUCCAUA ---...((((((.(((....(((((((((..(((((((((.((.(.(((......))).).)).))))))))))))).)))))...))).)).)))).. ( -35.60) >DroSim_CAF1 14999 97 - 1 --UUUUUGGAAGAGGAAUCCCCUUCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCGAUACCAGGAGGCUGUCUGCUAUCCAUA --....((((((.(((....(((((((((..(((((((((.((.(.(((......))).).)).))))))))))))).)))))...))).)).)))).. ( -35.60) >DroEre_CAF1 15579 96 - 1 --UUUUCGGAAGAGGAAUCCCC-UCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAAACUGUCUGCUCUCCAUA --.....(((.((((.....))-))((((.((((((((((.((.(.(((......))).).)).))))))))))..(((....)))..)))).)))... ( -34.60) >DroYak_CAF1 14150 97 - 1 --UUUUCGGAAGAGGAAUCCCCUUCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCGCUCCAUA --......((((.((....))))))((((..(((((((((.((.(.(((......))).).)).))))))))))))).((((((.....)).))))... ( -37.50) >consensus __UUUUUGGAAGAGGAAUCCCCUUCGGUAAAUUGAAUUGAAGCGUCGCCAGAAUGGGCAAGGCAUCAAUUCAAUACCAGGAGGCUGUCUGCUCUCCAUA .......(((((.(((....(((((((((..(((((((((.((.(.(((......))).).)).))))))))))))).)))))...))).)).)))... (-32.52 = -33.08 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:26 2006