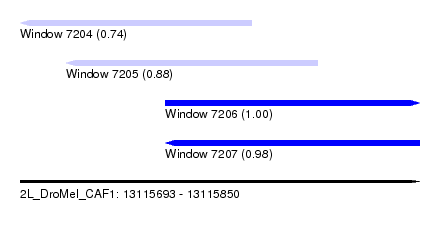
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,115,693 – 13,115,850 |
| Length | 157 |
| Max. P | 0.999530 |

| Location | 13,115,693 – 13,115,784 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.69 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13115693 91 - 22407834 AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUCCAGCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAACUCGCGU .......(((((.......(....).....(.(((.(((((((....))).))))(((((..........)))))))).)....))))).. ( -20.40) >DroSec_CAF1 8642 91 - 1 AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUCCAGCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAACUCGCGU .......(((((.......(....).....(.(((.(((((((....))).))))(((((..........)))))))).)....))))).. ( -20.40) >DroSim_CAF1 8842 91 - 1 AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUCCAGCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAACUCGCGU .......(((((.......(....).....(.(((.(((((((....))).))))(((((..........)))))))).)....))))).. ( -20.40) >DroEre_CAF1 10648 91 - 1 AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUCCAUCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAAUUCGCGC .((((((((((((......(....).......))..))))(((....)))....))))))...........(((((((......))))))) ( -17.92) >DroYak_CAF1 8949 91 - 1 AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUACAGCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAAAUCGCGC .(((((((((.........(....)....((((((...))))))...)))....))))))...........((((((........)))))) ( -16.20) >consensus AAAUGCAGCGAGAAAAGAGAGAAAUAAAAACGUCCAGCGUGCGUAUACGUAAUGUGCAUUAACAUCAACAAGUGCGGAGGAAAACUCGCGU .......(((((.......(....).....(.(((.(((((((....))).))))(((((..........)))))))).)....))))).. (-16.74 = -17.22 + 0.48)

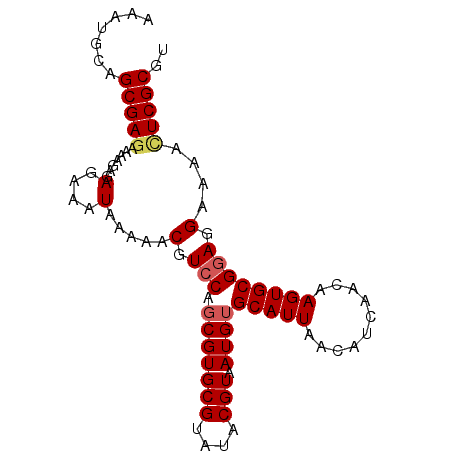
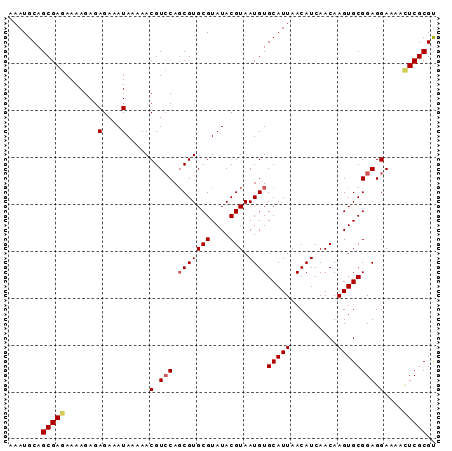
| Location | 13,115,711 – 13,115,810 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -11.27 |
| Energy contribution | -13.32 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13115711 99 - 22407834 CUGCAUAUCGCAAAAUG--------------CAAAAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-CCAGCGUGCGUAUACGUAAUGUGCAUUAACAUC-AACAAGU ((((((.(((((.....--------------.....))))).)))))).............-----..........(.-.((...((((....)))).))..).........-....... ( -19.90) >DroSec_CAF1 8660 106 - 1 CUGCAUAUCGCAAAAUGCAAAAU-------GCAAAAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-CCAGCGUGCGUAUACGUAAUGUGCAUUAACAUC-AACAAGU ((((((.(((((...(((.....-------)))...))))).)))))).............-----..........(.-.((...((((....)))).))..).........-....... ( -24.40) >DroSim_CAF1 8860 106 - 1 CUGCAUAUCGCAAAAUGCAAAAU-------GCAAAAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-CCAGCGUGCGUAUACGUAAUGUGCAUUAACAUC-AACAAGU ((((((.(((((...(((.....-------)))...))))).)))))).............-----..........(.-.((...((((....)))).))..).........-....... ( -24.40) >DroEre_CAF1 10666 92 - 1 CUGCAUAUCGCCA---------------------AAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-CCAUCGUGCGUAUACGUAAUGUGCAUUAACAUC-AACAAGU ((((((.((((..---------------------...)))).)))))).............-----..........(.-.(((..((((....)))))))..).........-....... ( -18.40) >DroWil_CAF1 10008 103 - 1 -----AACCACAAAAAUGUGAAU-------GCACAAAUAGAAA-----CAAGAACAAAAAAAAAGUAUAUAAAAAAAUACCAAUGUGCGUAUACGUAAUGUGCAUUAACAUCCAACAAGU -----..........((((.(((-------(((((........-----................((((........)))).....((((....)))).)))))))).))))......... ( -16.70) >DroYak_CAF1 8967 113 - 1 CUGCAAAUCGCAAAAUGCAAAAUGCAUAAUGCAAAAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-ACAGCGUGCGUAUACGUAAUGUGCAUUAACAUC-AACAAGU (((((..(((((...((((..........))))...)))))..))))).............-----..........((-(((...((((....)))).))))).........-....... ( -25.70) >consensus CUGCAUAUCGCAAAAUGCAAAAU_______GCAAAAUGCGAAAUGCAGCGAGAAAAGAGAG_____AAAUAAAAACGU_CCAGCGUGCGUAUACGUAAUGUGCAUUAACAUC_AACAAGU ((((((.(((((...(((............)))...))))).))))))....................................(..(((.......)))..)................. (-11.27 = -13.32 + 2.04)

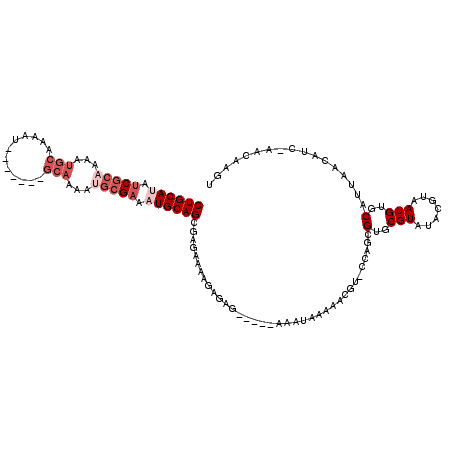
| Location | 13,115,750 – 13,115,850 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.11 |
| Mean single sequence MFE | -19.81 |
| Consensus MFE | -9.73 |
| Energy contribution | -11.38 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13115750 100 + 22407834 G-ACGUUUUUAUUU-----CUCUCUUUUCUCGCUGCAUUUCGCAUUUUG-------CAUUUUGCGAUAUGCAGUUCGCCGUCUGCCGCAUAAUUCUUAUUUUAAUGCUGCACU (-(((.........-----............(((((((.(((((.....-------.....))))).)))))))....))))(((.((((((........)).)))).))).. ( -23.41) >DroEre_CAF1 10705 93 + 1 G-ACGUUUUUAUUU-----CUCUCUUUUCUCGCUGCAUUUCGCAUU--------------UGGCGAUAUGCAGCUCGCCGUCUGCCGCAUAAUUCUUAUUUUAAUGCUGCACU (-(((.........-----............(((((((.((((...--------------..)))).)))))))....))))(((.((((((........)).)))).))).. ( -24.11) >DroWil_CAF1 10048 100 + 1 GUAUUUUUUUAUAUACUUUUUUUUUGUUCUUG-----UUUCUAUUUGUGCAUUCACAUUUUUGUGGUU-----UUUGCUG-AUGCCGCAUAAUUUAUAUUUUAAUGCCAAA-- (((((....((((...................-----........((((....))))....((((((.-----((....)-).)))))).....))))....)))))....-- ( -11.90) >consensus G_ACGUUUUUAUUU_____CUCUCUUUUCUCGCUGCAUUUCGCAUU_UG_________UUUUGCGAUAUGCAGUUCGCCGUCUGCCGCAUAAUUCUUAUUUUAAUGCUGCACU ...............................(((((((.((((...................)))).)))))))........(((.((((((........)).)))).))).. ( -9.73 = -11.38 + 1.64)

| Location | 13,115,750 – 13,115,850 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 64.11 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -7.83 |
| Energy contribution | -9.70 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.978018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13115750 100 - 22407834 AGUGCAGCAUUAAAAUAAGAAUUAUGCGGCAGACGGCGAACUGCAUAUCGCAAAAUG-------CAAAAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-C ..(((.((((.............)))).)))((((.....((((((.(((((.....-------.....))))).))))))...........(.-----...).....)))-) ( -22.82) >DroEre_CAF1 10705 93 - 1 AGUGCAGCAUUAAAAUAAGAAUUAUGCGGCAGACGGCGAGCUGCAUAUCGCCA--------------AAUGCGAAAUGCAGCGAGAAAAGAGAG-----AAAUAAAAACGU-C ..(((.((((.............)))).)))((((.(..(((((((.((((..--------------...)))).)))))))..).......(.-----...).....)))-) ( -26.62) >DroWil_CAF1 10048 100 - 1 --UUUGGCAUUAAAAUAUAAAUUAUGCGGCAU-CAGCAAA-----AACCACAAAAAUGUGAAUGCACAAAUAGAAA-----CAAGAACAAAAAAAAAGUAUAUAAAAAAAUAC --(((((((((.............(((.....-..)))..-----...((((....))))))))).))))......-----................................ ( -11.90) >consensus AGUGCAGCAUUAAAAUAAGAAUUAUGCGGCAGACGGCGAACUGCAUAUCGCAAAA_________CA_AAUGCGAAAUGCAGCGAGAAAAGAGAG_____AAAUAAAAACGU_C ..(((.((((.............)))).))).........((((((.((((...................)))).))))))................................ ( -7.83 = -9.70 + 1.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:22 2006