| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,113,953 – 13,114,072 |
| Length | 119 |
| Max. P | 0.940297 |

| Location | 13,113,953 – 13,114,072 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -15.11 |
| Energy contribution | -17.20 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13113953 119 + 22407834 CCUUGAAGCUACCUAAACAAAAUUGGCUCGACCCAAAAGUCACAAACAAUUUGAGUUUCCAAGGGGGAAACUCUAGUAAGUGAGGCAGUCAGCUUCAAGGCAAAUUUUUCACAAUGC-GA ((((((((((..((........((((......))))..(((.((....(((.((((((((.....)))))))).)))...)).)))))..)))))))))).................-.. ( -34.50) >DroSec_CAF1 7038 119 + 1 CCUUGAAGCUACCUAAACAAAAUUGGCUCGGCCCAAAAGUCACAAACAAUUUGAGUUUCCAAGGGUGAAACUCAAGUAAGUGAGGCAGUCAGCUUCAAGGCAAAUUUUUCACAAUGC-GA ((((((((((..((........((((......))))..(((.((....((((((((((((....).)))))))))))...)).)))))..)))))))))).................-.. ( -34.80) >DroSim_CAF1 7157 118 + 1 CCUUGAAGCUACCUAAAUAAAAUUGGCUCGAC-CAAAAGUCACAAACAAUUUGAGUUUCCAAGGGUGAAACUCAAGUAAGUGAGGCAGUCAGCUUCAAGGCAAAUUUUUCACAAUGC-GA ((((((((((..((........((((.....)-)))..(((.((....((((((((((((....).)))))))))))...)).)))))..)))))))))).................-.. ( -35.00) >DroEre_CAF1 9050 107 + 1 C-----------CUAAACAAAAUUGGCUCGACCCAAAAGUCGCAAACAACUUGAGUUUCCAAGG-GGAAACUCAACUAACUGAGGCAGUCAGCCUCAAGGCAAAUUCUUCACAAUGC-GA .-----------..........((((......))))...(((((......((((((((((....-)))))))))).....((((((.....)))))).................)))-)) ( -30.90) >DroYak_CAF1 7258 119 + 1 CCUCCAAGCUACCUAAACAAAAUUGGCUUUACCCAAAAGUCACACACAAUUUGAGUUUCCAAGGGGGAAACUGAAGAAACUCAGGCAGUCAGCUUCAAGGCAAAUUUUUCACGAAGC-GA (((..(((((..((.........(((((((.....))))))).......((((((((((.....((....))...)))))))))).))..)))))..))).................-.. ( -26.50) >DroAna_CAF1 5513 98 + 1 ---------UUCCU-CGCCAAAUUGGCUCAACCCAAAAGUCAAAAUCAAUU--GGUUUCCAGAG----------AGUGGCCGAAACAGCCAAUGGCGAUGCGAAUUUUUCACAAUGCUGG ---------(((((-(((((..((((((..........((((...((..((--((...)))).)----------).))))......)))))))))))).).)))................ ( -20.79) >consensus CCUUGAAGCUACCUAAACAAAAUUGGCUCGACCCAAAAGUCACAAACAAUUUGAGUUUCCAAGGGGGAAACUCAAGUAACUGAGGCAGUCAGCUUCAAGGCAAAUUUUUCACAAUGC_GA ......................((((......))))..(((.......(((((((((((.......)))))))))))...((((((.....)))))).)))................... (-15.11 = -17.20 + 2.09)

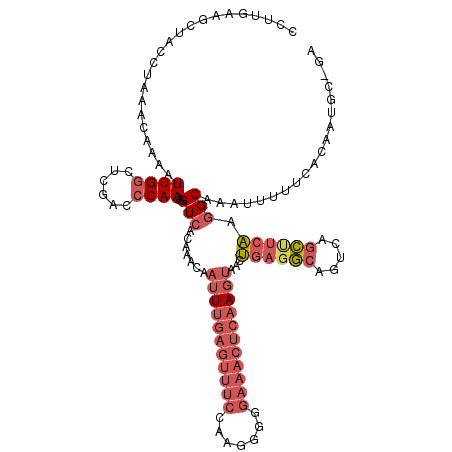

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:18 2006