| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,113,257 – 13,113,394 |
| Length | 137 |
| Max. P | 0.726492 |

| Location | 13,113,257 – 13,113,363 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.12 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13113257 106 - 22407834 AAUUUCUUUUUAUCUUAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAAAGAAAACUACCCUGAU------ACUCUUACACGCUUAACAGGCCAUGUUACAACUGGGACCAA ..............(((((((........))))))).((((.(((....))).)))).(((....------......(((.(((.....)))..))).......)))..... ( -14.93) >DroSec_CAF1 6330 104 - 1 AAU--CUAUUUAUCUUAAGGACAAAAUCAUCCUUAACAGUUCUCUUAAGAGAGAACUACCCUGGU------ACUCUUGCAUCCCUAACAGGCCCUGUUAAAACUGGGACCAA ...--.....(((((((((((........))))))).((((((((....)))))))).....)))------).................(((((.((....)).))).)).. ( -24.00) >DroSim_CAF1 6444 104 - 1 AAU--CUCUUUAUCUGAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAGGGAGAACUACCCUGGU------ACUCUUACACCCCUAACAGGACCUGUUAAAACUGGGACCAA ...--...........(((((........)))))...((((((((....)))))))).(((.(((------........)))..((((((...)))))).....)))..... ( -22.70) >DroEre_CAF1 8350 109 - 1 AAU--CUCUUUAUCUCAAACACAAUACCUUCUCUGGCAGUUCUCUUAAGGGAUAACUACCCUAGUGCUCUUACUCUUACGUCCUUAACAGGCCAUGUUAAAAC-GGGACCAA ...--.....................((......))..((.....((((((...((((...)))).)))))).....))((((((((((.....)))))....-)))))... ( -16.30) >DroYak_CAF1 6560 104 - 1 AAU--CUCUUUAUCUUAGGGACAAUAUAUUCGUUGACAGUUCUCUUAAGGGAGAACUACCCCAGU------ACUCUUACACCCUUCACAGGCCAUGUUAAAACCGGGUCCAA ...--...........((((.((((......))))..((((((((....))))))))........------.........)))).....((((..((....))..))))... ( -20.00) >consensus AAU__CUCUUUAUCUUAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAGGGAGAACUACCCUGGU______ACUCUUACACCCUUAACAGGCCAUGUUAAAACUGGGACCAA ..............(((((((........))))))).((((((((....)))))))).(((.(((......)))..........(((((.....))))).....)))..... (-14.64 = -14.96 + 0.32)

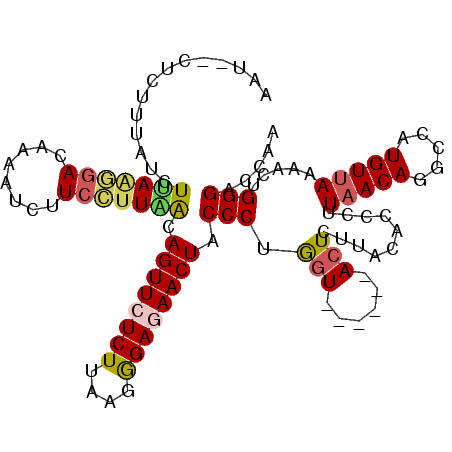
| Location | 13,113,292 – 13,113,394 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13113292 102 + 22407834 AAGAGU------AUCAGGGUAGUUUUCUUUUAAGAGAACUGUUAAGGAAGAUUUUGUCCUUAAGAUAAAAAGAAAUUUAAAAGUAUUCAUUGAAUUUAGGCUUAACAU ..((((------(((....(((((((((....)))))))))(((((((........))))))))))......((((((((.........))))))))..))))..... ( -18.80) >DroSec_CAF1 6365 100 + 1 AAGAGU------ACCAGGGUAGUUCUCUCUUAAGAGAACUGUUAAGGAUGAUUUUGUCCUUAAGAUAAAUAG--AUUUAAAAGAAUUCAUUGAAUUUAGGCUUAACAU ..((((------.......(((((((((....)))))))))((((((((......))))))))......(((--((((((.........))))))))).))))..... ( -23.10) >DroSim_CAF1 6479 100 + 1 AAGAGU------ACCAGGGUAGUUCUCCCUUAAGAGAACUGUUAAGGAAGAUUUUGUCCUUCAGAUAAAGAG--AUUAAAAAGAAAACAUUGAAUUUAGGCUCAACAU ..((((------.((..((((((((((......))))))))))..))....(((((((.....)))))))..--.........................))))..... ( -19.90) >DroEre_CAF1 8384 106 + 1 AAGAGUAAGAGCACUAGGGUAGUUAUCCCUUAAGAGAACUGCCAGAGAAGGUAUUGUGUUUGAGAUAAAGAG--AUUAAAAAGAAUUCAUUGAAUUCGGGCUCAAUAU .............((..(((((((.((......)).)))))))..))...((((((.(((((((...((..(--(((......))))..))...))))))).)))))) ( -22.90) >DroYak_CAF1 6595 100 + 1 AAGAGU------ACUGGGGUAGUUCUCCCUUAAGAGAACUGUCAACGAAUAUAUUGUCCCUAAGAUAAAGAG--AUUAAAAAGAAGUCAUUGAGCUCAGGCUCAACAU ..((((------.(((((.((((((((......))))))))((((........(((((.....)))))...(--(((.......)))).)))).)))))))))..... ( -24.50) >consensus AAGAGU______ACCAGGGUAGUUCUCCCUUAAGAGAACUGUUAAGGAAGAUUUUGUCCUUAAGAUAAAGAG__AUUAAAAAGAAUUCAUUGAAUUUAGGCUCAACAU ..((((.......((..((((((((((......))))))))))..))....(((((((.....))))))).............................))))..... (-15.48 = -15.40 + -0.08)

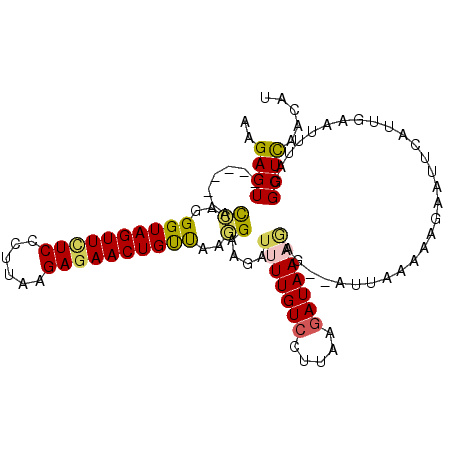
| Location | 13,113,292 – 13,113,394 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -11.66 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13113292 102 - 22407834 AUGUUAAGCCUAAAUUCAAUGAAUACUUUUAAAUUUCUUUUUAUCUUAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAAAGAAAACUACCCUGAU------ACUCUU .........................................(((((((((((........))))))).((((.(((....))).)))).....)))------)..... ( -10.40) >DroSec_CAF1 6365 100 - 1 AUGUUAAGCCUAAAUUCAAUGAAUUCUUUUAAAU--CUAUUUAUCUUAAGGACAAAAUCAUCCUUAACAGUUCUCUUAAGAGAGAACUACCCUGGU------ACUCUU .......(((........................--.........(((((((........))))))).((((((((....)))))))).....)))------...... ( -18.00) >DroSim_CAF1 6479 100 - 1 AUGUUGAGCCUAAAUUCAAUGUUUUCUUUUUAAU--CUCUUUAUCUGAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAGGGAGAACUACCCUGGU------ACUCUU ((((((((......))))))))............--.....((((..(((((........)))))...((((((((....)))))))).....)))------)..... ( -17.40) >DroEre_CAF1 8384 106 - 1 AUAUUGAGCCCGAAUUCAAUGAAUUCUUUUUAAU--CUCUUUAUCUCAAACACAAUACCUUCUCUGGCAGUUCUCUUAAGGGAUAACUACCCUAGUGCUCUUACUCUU .....((((.(((((((...))))))........--.............................((.((((.(((....))).)))).))...).))))........ ( -15.40) >DroYak_CAF1 6595 100 - 1 AUGUUGAGCCUGAGCUCAAUGACUUCUUUUUAAU--CUCUUUAUCUUAGGGACAAUAUAUUCGUUGACAGUUCUCUUAAGGGAGAACUACCCCAGU------ACUCUU ..(((((((....)))))))..............--............(((.((((......))))..((((((((....))))))))..)))...------...... ( -24.00) >consensus AUGUUGAGCCUAAAUUCAAUGAAUUCUUUUUAAU__CUCUUUAUCUUAAGGACAAAAUCUUCCUUAACAGUUCUCUUAAGGGAGAACUACCCUGGU______ACUCUU .(((((((......)))))))........................(((((((........))))))).((((((((....)))))))).................... (-11.66 = -11.90 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:17 2006