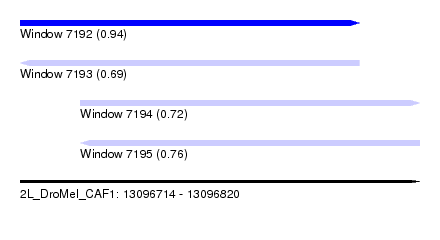
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,096,714 – 13,096,820 |
| Length | 106 |
| Max. P | 0.937333 |

| Location | 13,096,714 – 13,096,804 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 63.58 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -5.78 |
| Energy contribution | -6.67 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13096714 90 + 22407834 UUUCGAUUA------AA-UAUUG-----------GCUUGUUUUGCUCUUGUAAUCACUACAAAUGAUGAUCAUGCGAUCAUCAUUUUACUGACUGAUCUAUCUCACCA ....(((((------.(-((..(-----------((.......)))..)))..(((.((.(((((((((((....))))))))))))).))).))))).......... ( -20.10) >DroSec_CAF1 77831 90 + 1 UUUAGAUUG------AA-UAUUG-----------GCUUGUUUUGCUCUUGCAAUCACUAAAAAUGAUGAUCAUGAGAUCAUCAUUUUACAGCCUGAUCUAUCUCACCA ...((((.(------(.-....(-----------(((.((.((((....))))......((((((((((((....))))))))))))))))))...)).))))..... ( -24.80) >DroSim_CAF1 75690 90 + 1 UUUAGAUUG------AA-UAUUG-----------GCUUGUCUUGCUUUUGCAAUCACUAAAAAUGAUGAUCAUGCGAUCAUCAAAUUACUGCCUGAUCUAUCUCACCA ...((((.(------(.-....(-----------((.....((((....))))..........((((((((....)))))))).......)))...)).))))..... ( -19.80) >DroEre_CAF1 78955 87 + 1 UUUGCUUUG------AA-UAUUACGUUACAACAAGCUUGUUGUGAUUUUACA--------------UGAUCACGCGAUCAUCAUUUCGGUGCCUGAUCUAUCUCACCA ........(------((-......(((((((((....))))))))).....(--------------(((((....))))))...)))((((...((....)).)))). ( -18.10) >DroYak_CAF1 78670 103 + 1 UUUAGUUUGUAUACGAG-UAUUACAUUACAA----CUUGUUGUGCGUUUACAAUCACUAAAAAUGAUGAUCAUGCGAUCACCAAUUCGCUGCCUGAUCUAUCUCACCA (((((((((((.((..(-(((.(((......----..))).)))))).)))))..))))))...((((((((.((((........))))....))))).)))...... ( -22.70) >DroAna_CAF1 63668 72 + 1 UUUCCAUGA------AAACCUUU-----------CCUUAUCGUU----------CCUUAGAAAU-AUCAACUUGUGUUUAUUUUUUCA--GA------AAUCUGAUCG .......((------........-----------..........----------....((((((-(..(((....))))))))))(((--(.------...)))))). ( -5.10) >consensus UUUAGAUUG______AA_UAUUG___________GCUUGUUGUGCUUUUGCAAUCACUAAAAAUGAUGAUCAUGCGAUCAUCAUUUCACUGCCUGAUCUAUCUCACCA ...............................................................((((((((....))))))))......................... ( -5.78 = -6.67 + 0.89)

| Location | 13,096,714 – 13,096,804 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 63.58 |
| Mean single sequence MFE | -19.11 |
| Consensus MFE | -6.17 |
| Energy contribution | -6.78 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13096714 90 - 22407834 UGGUGAGAUAGAUCAGUCAGUAAAAUGAUGAUCGCAUGAUCAUCAUUUGUAGUGAUUACAAGAGCAAAACAAGC-----------CAAUA-UU------UAAUCGAAA ((((((......))(((((.(((((((((((((....))))))))))).)).)))))...............))-----------))...-..------......... ( -21.70) >DroSec_CAF1 77831 90 - 1 UGGUGAGAUAGAUCAGGCUGUAAAAUGAUGAUCUCAUGAUCAUCAUUUUUAGUGAUUGCAAGAGCAAAACAAGC-----------CAAUA-UU------CAAUCUAAA .....((((.((...((((..((((((((((((....))))))))))))..((..((((....)))).)).)))-----------)....-.)------).))))... ( -26.40) >DroSim_CAF1 75690 90 - 1 UGGUGAGAUAGAUCAGGCAGUAAUUUGAUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGCAAAAGCAAGACAAGC-----------CAAUA-UU------CAAUCUAAA .....((((.((...(((..(((..((((((((....))))))))...)))((..((((....)))).))..))-----------)....-.)------).))))... ( -21.80) >DroEre_CAF1 78955 87 - 1 UGGUGAGAUAGAUCAGGCACCGAAAUGAUGAUCGCGUGAUCA--------------UGUAAAAUCACAACAAGCUUGUUGUAACGUAAUA-UU------CAAAGCAAA ..((((.((((((((.((...............)).))))).--------------)))....)))).....((((...(((......))-).------..))))... ( -17.86) >DroYak_CAF1 78670 103 - 1 UGGUGAGAUAGAUCAGGCAGCGAAUUGGUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGUAAACGCACAACAAG----UUGUAAUGUAAUA-CUCGUAUACAAACUAAA ....(((.((...((.((((((((.((((((((....)))))))).)))..(((........))).......)----))))..))...))-))).............. ( -19.60) >DroAna_CAF1 63668 72 - 1 CGAUCAGAUU------UC--UGAAAAAAUAAACACAAGUUGAU-AUUUCUAAGG----------AACGAUAAGG-----------AAAGGUUU------UCAUGGAAA ...((((...------.)--)))...........((.(..(((-.(((((....----------........))-----------))).))).------.).)).... ( -7.30) >consensus UGGUGAGAUAGAUCAGGCAGUAAAAUGAUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGCAAAAGCAAAACAAGC___________CAAUA_UU______CAAUCUAAA .........................((((((((....))))))))............................................................... ( -6.17 = -6.78 + 0.61)


| Location | 13,096,730 – 13,096,820 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -16.56 |
| Consensus MFE | -7.56 |
| Energy contribution | -9.00 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13096730 90 + 22407834 ------GCUUGUUUUGCUCUUGUAAUCACUACAAAUGAUGAUCAUGCGAUCAUCAUUUUACUGACUGAUCUAUCUCACCAAUUAGUUAAAUAAUCA ------(.((((((.(((.(((...(((.((.(((((((((((....))))))))))))).))).(((......))).)))..))).)))))).). ( -17.60) >DroSec_CAF1 77847 90 + 1 ------GCUUGUUUUGCUCUUGCAAUCACUAAAAAUGAUGAUCAUGAGAUCAUCAUUUUACAGCCUGAUCUAUCUCACCAAUUAGUUAAAUAAUCA ------(((.((.((((....))))......((((((((((((....)))))))))))))))))................................ ( -18.90) >DroSim_CAF1 75706 90 + 1 ------GCUUGUCUUGCUUUUGCAAUCACUAAAAAUGAUGAUCAUGCGAUCAUCAAAUUACUGCCUGAUCUAUCUCACCAAUUAGUUAAAUAAUCA ------.......((((....))))..(((((...((((((((....))))))))..........(((......)))....))))).......... ( -15.00) >DroEre_CAF1 78976 82 + 1 CAACAAGCUUGUUGUGAUUUUACA--------------UGAUCACGCGAUCAUCAUUUCGGUGCCUGAUCUAUCUCACCAAUUAGCUAAAUAAUCA .....((((.(((((((..(....--------------.(((((.((.(((........))))).))))).)..))).)))).))))......... ( -16.40) >DroYak_CAF1 78697 92 + 1 CAA----CUUGUUGUGCGUUUACAAUCACUAAAAAUGAUGAUCAUGCGAUCACCAAUUCGCUGCCUGAUCUAUCUCACCAAUUAGUUAAAUAAUCA .((----((.((((((.(......((((.......))))(((((.((((........))))....)))))...).)).)))).))))......... ( -14.90) >consensus ______GCUUGUUUUGCUUUUGCAAUCACUAAAAAUGAUGAUCAUGCGAUCAUCAUUUUACUGCCUGAUCUAUCUCACCAAUUAGUUAAAUAAUCA ........((((((.((..................((((((((....))))))))..........(((......))).......)).))))))... ( -7.56 = -9.00 + 1.44)



| Location | 13,096,730 – 13,096,820 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -20.69 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13096730 90 - 22407834 UGAUUAUUUAACUAAUUGGUGAGAUAGAUCAGUCAGUAAAAUGAUGAUCGCAUGAUCAUCAUUUGUAGUGAUUACAAGAGCAAAACAAGC------ .((((((((.(((....))).)))).))))(((((.(((((((((((((....))))))))))).)).))))).................------ ( -22.00) >DroSec_CAF1 77847 90 - 1 UGAUUAUUUAACUAAUUGGUGAGAUAGAUCAGGCUGUAAAAUGAUGAUCUCAUGAUCAUCAUUUUUAGUGAUUGCAAGAGCAAAACAAGC------ (((((((((.(((....))).)))).))))).((((.((((((((((((....))))))))))))))))..((((....)))).......------ ( -25.30) >DroSim_CAF1 75706 90 - 1 UGAUUAUUUAACUAAUUGGUGAGAUAGAUCAGGCAGUAAUUUGAUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGCAAAAGCAAGACAAGC------ (((((((((.(((....))).)))).)))))..((.(((..((((((((....))))))))...))).)).((((....)))).......------ ( -20.60) >DroEre_CAF1 78976 82 - 1 UGAUUAUUUAGCUAAUUGGUGAGAUAGAUCAGGCACCGAAAUGAUGAUCGCGUGAUCA--------------UGUAAAAUCACAACAAGCUUGUUG .........((((.....((((.((((((((.((...............)).))))).--------------)))....))))....))))..... ( -16.76) >DroYak_CAF1 78697 92 - 1 UGAUUAUUUAACUAAUUGGUGAGAUAGAUCAGGCAGCGAAUUGGUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGUAAACGCACAACAAG----UUG ........(((((..((((((.....(((((......(((.((((((((....)))))))).)))...))))).....))).)))..))----))) ( -18.80) >consensus UGAUUAUUUAACUAAUUGGUGAGAUAGAUCAGGCAGUAAAAUGAUGAUCGCAUGAUCAUCAUUUUUAGUGAUUGCAAAAGCAAAACAAGC______ (((((((((.(((....))).)))).)))))..........((((((((....))))))))................................... (-12.86 = -13.18 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:11 2006