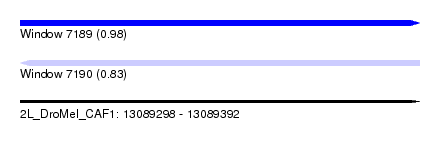
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,089,298 – 13,089,392 |
| Length | 94 |
| Max. P | 0.981694 |

| Location | 13,089,298 – 13,089,392 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.05 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
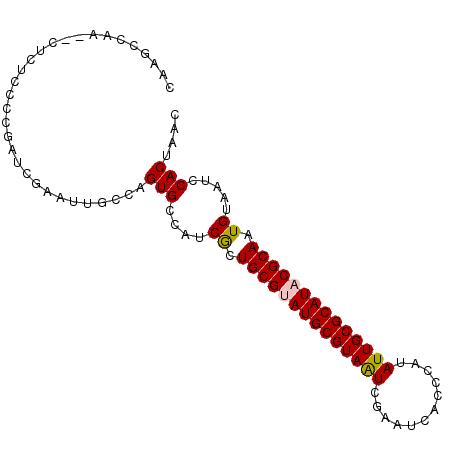
>2L_DroMel_CAF1 13089298 94 + 22407834 CAAGCCAA--CUCUCCCCGAUCGAAUUGCCAGUGCCAUCGCUGCGGAUGCGUAAUCGAAUCAUCGAUAUUGCGCAUACGCAAUGUAAUCCACUUCC ........--........(((...((((((((((....))))).(.((((((((((((....)))))..))))))).))))))...)))....... ( -22.60) >DroPse_CAF1 91626 81 + 1 CAGCCAACAU---------------GUUUCAGUGUCAGCGCUGCGUAUGCGUAGUCGAACGUCUCAUAUUGCGCAUACGCAAUGUAAUCCACAAAC ..((..((((---------------......))))..))..((((((((((((((.((.....))..))))))))))))))............... ( -25.50) >DroSec_CAF1 70435 94 + 1 CAAGCCAA--CUCUACACGAUCGAUUUGCCAGUGCCAUCACUGCGUAUGCGUAAUCGAAUCAUCCAUAUUGCGCAUACGCAAUGAAAUCCACUCGC ........--.................((.((((...(((.((((((((((((((.((....))...)))))))))))))).)))....)))).)) ( -27.70) >DroSim_CAF1 68122 94 + 1 UAAGCCAA--CUCUCCCCGAUCGAAUUGCCAGUGCCACCACUGCGUAUGCGUAAUCGAAUCAUCCAUAUUGCGCAUACGCAAUGCAAUCCACUUCC ........--............(.(((((.((((....))))(((((((((((((.((....))...)))))))))))))...))))).)...... ( -25.80) >DroYak_CAF1 71095 96 + 1 CAAUCCACAUAUUUCCCCCUUCGAAUUGCCAGUGACAUCGCUGCGGAUGCGUAAUCGAAUCACCCAUAUUGCGCAUACGCAAUGCAAUCCACUCGC ......................(.(((((.((((....))))(((.(((((((((............))))))))).)))...))))).)...... ( -21.80) >DroPer_CAF1 88328 81 + 1 CAGCCAACAU---------------GUUUCAGUGUCAGCGCUGCGUAUGCGUAGUCGAACGUCUCAUAUUGCGCAUACGCAAUGUAAUCCACAAAC ..((..((((---------------......))))..))..((((((((((((((.((.....))..))))))))))))))............... ( -25.50) >consensus CAAGCCAA__CUCUCCCCGAUCGAAUUGCCAGUGCCAUCGCUGCGUAUGCGUAAUCGAAUCACCCAUAUUGCGCAUACGCAAUGUAAUCCACUAAC ...............................(((....((.((((((((((((((............)))))))))))))).)).....))).... (-21.16 = -21.05 + -0.11)

| Location | 13,089,298 – 13,089,392 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -17.43 |
| Energy contribution | -18.57 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
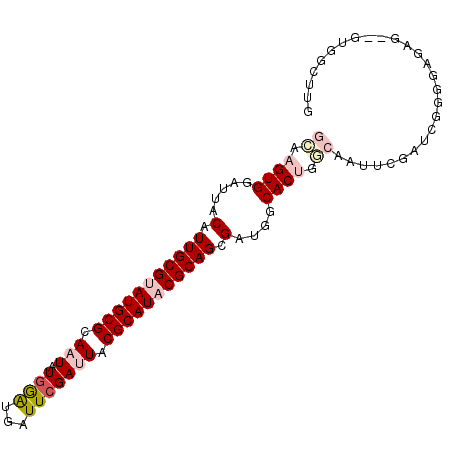
>2L_DroMel_CAF1 13089298 94 - 22407834 GGAAGUGGAUUACAUUGCGUAUGCGCAAUAUCGAUGAUUCGAUUACGCAUCCGCAGCGAUGGCACUGGCAAUUCGAUCGGGGAGAG--UUGGCUUG ..((((.((((...(((((.(((((....(((((....)))))..))))).)))))(((..((....))...))).........))--)).)))). ( -27.40) >DroPse_CAF1 91626 81 - 1 GUUUGUGGAUUACAUUGCGUAUGCGCAAUAUGAGACGUUCGACUACGCAUACGCAGCGCUGACACUGAAAC---------------AUGUUGGCUG ..............(((((((((((.....((((...))))....))))))))))).((..(((.......---------------.)))..)).. ( -23.10) >DroSec_CAF1 70435 94 - 1 GCGAGUGGAUUUCAUUGCGUAUGCGCAAUAUGGAUGAUUCGAUUACGCAUACGCAGUGAUGGCACUGGCAAAUCGAUCGUGUAGAG--UUGGCUUG ((.((((....((((((((((((((....((.((....)).))..))))))))))))))...)))).))((..(((((.....)).--)))..)). ( -34.90) >DroSim_CAF1 68122 94 - 1 GGAAGUGGAUUGCAUUGCGUAUGCGCAAUAUGGAUGAUUCGAUUACGCAUACGCAGUGGUGGCACUGGCAAUUCGAUCGGGGAGAG--UUGGCUUA ...((((.....(((((((((((((....((.((....)).))..)))))))))))))....))))(.((((((.........)))--))).)... ( -32.00) >DroYak_CAF1 71095 96 - 1 GCGAGUGGAUUGCAUUGCGUAUGCGCAAUAUGGGUGAUUCGAUUACGCAUCCGCAGCGAUGUCACUGGCAAUUCGAAGGGGGAAAUAUGUGGAUUG ((.((((((((((((((((....))))))..(((((...........)))))...))))).))))).))..(((....)))............... ( -28.80) >DroPer_CAF1 88328 81 - 1 GUUUGUGGAUUACAUUGCGUAUGCGCAAUAUGAGACGUUCGACUACGCAUACGCAGCGCUGACACUGAAAC---------------AUGUUGGCUG ..............(((((((((((.....((((...))))....))))))))))).((..(((.......---------------.)))..)).. ( -23.10) >consensus GCAAGUGGAUUACAUUGCGUAUGCGCAAUAUGGAUGAUUCGAUUACGCAUACGCAGCGAUGGCACUGGCAAUUCGAUCGGGGAGAG__GUGGCUUG ((.((((.....(.(((((((((((.(((.((((...))))))).))))))))))).)....)))).))........................... (-17.43 = -18.57 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:07 2006