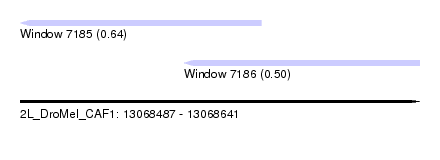
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,068,487 – 13,068,641 |
| Length | 154 |
| Max. P | 0.642467 |

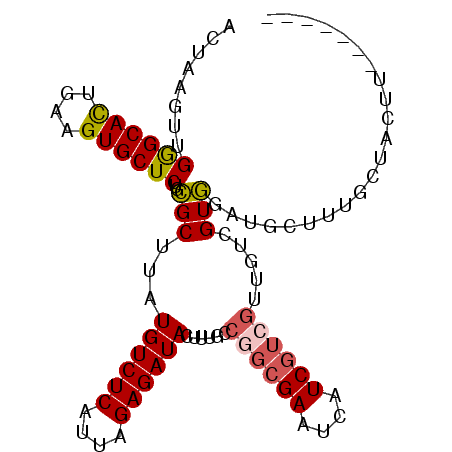
| Location | 13,068,487 – 13,068,580 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.63 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13068487 93 - 22407834 ACUAAGUUGGGCACUGAAGUGCUCUGCGCUUAUGUCUCAUUAGAGAUACUUUGCCGGCGAAUCAUCGUCGUUGGCGUGGAUGCUUUGCUACUU------- ..(((((.((((((....))))))...)))))((((((....))))))....((((((((.......))))))))((((........))))..------- ( -31.70) >DroPse_CAF1 65828 75 - 1 ACUUAGUUGAGCAUUGAAGUGCUCUGUGCUUAUGUCUCAUUAGAGAUACUUUACCAGCGAAUCAUCG------UCGUAGUC------------------- ....(((.((((((....))))))...)))..((((((....))))))..((((..((((....)))------).))))..------------------- ( -17.70) >DroSim_CAF1 45752 93 - 1 ACUAAGUUGGGCACUGAAGUGCUCUGCGCUUAUGUCUCAUUAGAGAUACUUUGCCGGCGAAUCAUCGUCGUUGGCGUGGAUGCUUUGCUACUU------- ..(((((.((((((....))))))...)))))((((((....))))))....((((((((.......))))))))((((........))))..------- ( -31.70) >DroEre_CAF1 49945 93 - 1 ACUAAGUUGGGCACUGAAGUGCUCUGCGCUUAUGUCUCAUUAGAGAUACUUUGCCGGCGAAUCAUCGUCGUUGUCGUGGAUGCUUUGCUACUU------- .....((.((((((....)))))).))((...((((((....))))))....)).((((((.((((..((....))..)))).))))))....------- ( -29.00) >DroYak_CAF1 49714 100 - 1 ACUAAGUUGGGCACUGAAGUGCUCUGCGCUUAUGUCUCAUUAGAGAUACUUUGCCGGCGAAUCAUCGUCGUUGUCGUGGAUGCUUUGCUACUUUGCUACU .....((.((((((....)))))).))((...((((((....))))))....)).((((((.((((..((....))..)))).))))))........... ( -29.00) >DroPer_CAF1 62601 81 - 1 ACUUAGUUGAGCAUUGAAGUGCUCUGUGCUUAUGUCUCAUUAGAGAUACUUUACCAGCGAAUCAUCAUCGUAGUCGUAGUC------------------- ....(((.((((((....))))))...)))..((((((....))))))........((((..(......)...))))....------------------- ( -17.00) >consensus ACUAAGUUGGGCACUGAAGUGCUCUGCGCUUAUGUCUCAUUAGAGAUACUUUGCCGGCGAAUCAUCGUCGUUGUCGUGGAUGCUUUGCUACUU_______ ........((((((....))))))..(((...((((((....))))))......((((((....)))))).....)))...................... (-19.69 = -19.63 + -0.05)

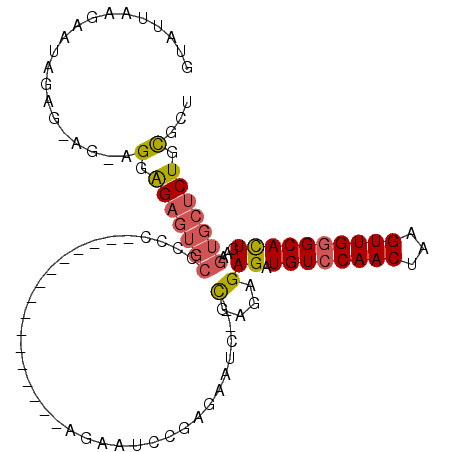
| Location | 13,068,550 – 13,068,641 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13068550 91 - 22407834 GUAUUAAGAAUAGAG-AGAAGGAGAGUGCCCUC----------------AGAAUCCGAGAAUC--UGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU ...............-....(.((((..(.(((----------------(((.((...)).))--))))..((.((((((((...))))))))))...)..)))).)... ( -28.30) >DroSec_CAF1 50061 91 - 1 GUAUUAAGAAUAGAA-AGCAGGGGAGUGCCCCC----------------AGAAGCCGAGAAUC--UGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU ...............-(((.((((.....))))----------------....((.(((...(--(.....((.((((((((...))))))))))..))..))).))))) ( -26.80) >DroSim_CAF1 45815 91 - 1 GUAUUAAGAAUAGAG-AGAAGGAGAGUGCCCCC----------------AGAAUCUGAGAAUC--UGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU ...............-....(.((((..(...(----------------((..(((.((...)--).)))....((((((((...)))))))))))..)..)))).)... ( -24.40) >DroEre_CAF1 50008 96 - 1 GUAUUAAGAUUAGAG-A---GGAGAGUGCCCCCGGAAU--------CCGAGAAUCCGAGAAUC--CGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU .............((-.---(.((((..(...((((.(--------(((......)).)).))--))....((.((((((((...))))))))))...)..)))).).)) ( -29.20) >DroYak_CAF1 49784 104 - 1 GUAUUAAGAAUUGAG-A---GGAGAGUGCCCCCUGAACUCGAGAACUCGAGAAUCCGAGAAUC--CGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU .............((-.---(.((((..(........((((.((.((((......))))..))--))))..((.((((((((...))))))))))...)..)))).).)) ( -31.20) >DroPer_CAF1 62652 83 - 1 GUAUCAAAAAAAGAGCAGUAGGGGC-------------------------GAAG--GAGGUUCGGCGAGAGAGAUGUCCAACUUAGUUGAGCAUUGAAGUGCUCUGUGCU ((((.......((((((.(...(.(-------------------------(((.--....)))).)......((((..((((...))))..))))..).)))))))))). ( -19.51) >consensus GUAUUAAGAAUAGAG_AG_AGGAGAGUGCCCCC________________AGAAUCCGAGAAUC__CGAGAGAGAUGUCCAACUAAGUUGGGCACUGAAGUGCUCUGCGCU ....................(.(((((((....................................(....)((.((((((((...))))))))))...))))))).)... (-15.86 = -16.12 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:03 2006