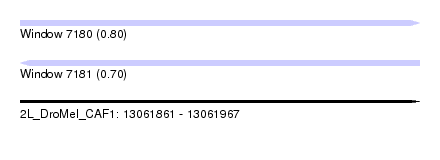
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,061,861 – 13,061,967 |
| Length | 106 |
| Max. P | 0.803503 |

| Location | 13,061,861 – 13,061,967 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -16.92 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
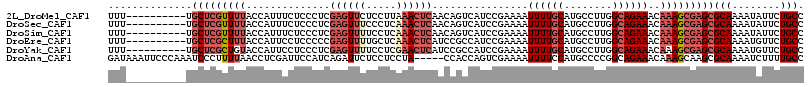
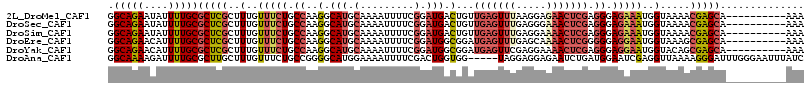
>2L_DroMel_CAF1 13061861 106 + 22407834 UUU----------UGCUCGUUUUACCAUUUCUCCCUCGAGUUCUCCUUAAACUCAACAGUCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUAUUCUGCC ...----------.(((((((((....(((((.((..(((((.......)))))...((.(((.(((.....))).))).)).)).))))).)))))))))(((........))). ( -23.80) >DroSec_CAF1 43484 106 + 1 UUU----------UGCUCGUUUUACCAUUUCUCCCUCGAGUUUCCCUCAAACUCAACAGUCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUAUUCUGCC ...----------.(((((((((....(((((.((..((((((.....))))))...((.(((.(((.....))).))).)).)).))))).)))))))))(((........))). ( -24.90) >DroSim_CAF1 39148 106 + 1 UUU----------UGCUCGUUUUACCAUUUCUCCCUCGAGUUUUCCUCAAACUCAACAGUCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUAUUCUGCC ...----------.(((((((((....(((((.((..((((((.....))))))...((.(((.(((.....))).))).)).)).))))).)))))))))(((........))). ( -24.90) >DroEre_CAF1 43216 106 + 1 UUU----------UGCUCGCUUUACCAUUCCUCCCCCGAGUUUUGCUCAAACUCAUCCGCCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUGUUCUGCC ...----------.(((((((((..............((((((.....)))))).((.((((....................)))).))...)))))))))(((........))). ( -26.95) >DroYak_CAF1 43063 106 + 1 UUU----------UGCUCGCUGUACCAUUCCUCCCUCGAGUUUUCCUCGAACUCAUCCGCCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUGUUCUGCC ...----------.(((((((................((((((.....)))))).((.((((....................)))).)).....)))))))(((........))). ( -25.55) >DroAna_CAF1 31810 111 + 1 GAUAAAUUCCCAAAUCCCUUUUAACCUCGAUUCCAUCAGAUUCUCCUCCUA-----CCACCAGUCGAAAAUUUUCCAUGCCCCGGCAGAAACAAAGCAAGCGCAAAAUCUUUUGCC ..........................((((((......((......))...-----.....))))))................(((((((.....((....))......))))))) ( -9.74) >consensus UUU__________UGCUCGUUUUACCAUUUCUCCCUCGAGUUUUCCUCAAACUCAACAGCCAUCCGAAAAUUUUGCAUGCCUUGGCAGAAACAAAGCGAGCGCAAAAUAUUCUGCC ..............(((((((((..............((((((.....))))))................((((((........))))))..)))))))))(((........))). (-16.92 = -18.78 + 1.86)
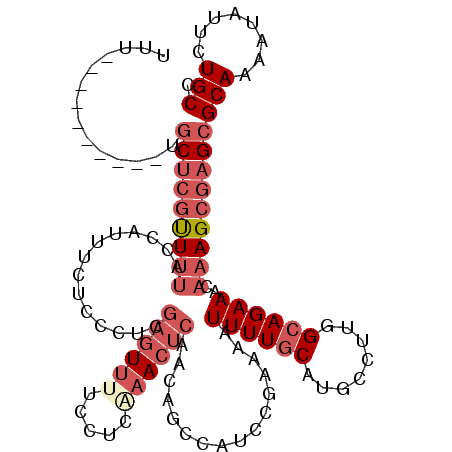
| Location | 13,061,861 – 13,061,967 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -20.79 |
| Energy contribution | -22.35 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
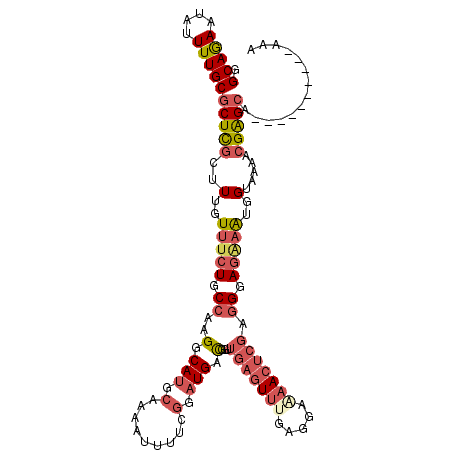
>2L_DroMel_CAF1 13061861 106 - 22407834 GGCAGAAUAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGACUGUUGAGUUUAAGGAGAACUCGAGGGAGAAAUGGUAAAACGAGCA----------AAA .(((((....)))))(((((..(..(((((.((..(((((.(.........).))).)).((((((((.....)))))))))).)))))..).....))))).----------... ( -31.30) >DroSec_CAF1 43484 106 - 1 GGCAGAAUAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGACUGUUGAGUUUGAGGGAAACUCGAGGGAGAAAUGGUAAAACGAGCA----------AAA .(((((....)))))(((((..(..(((((.((..(((((.(.........).))).)).((((((((.....)))))))))).)))))..).....))))).----------... ( -31.10) >DroSim_CAF1 39148 106 - 1 GGCAGAAUAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGACUGUUGAGUUUGAGGAAAACUCGAGGGAGAAAUGGUAAAACGAGCA----------AAA .(((((....)))))(((((..(..(((((.((..(((((.(.........).))).)).((((((((.....)))))))))).)))))..).....))))).----------... ( -31.80) >DroEre_CAF1 43216 106 - 1 GGCAGAACAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGGCGGAUGAGUUUGAGCAAAACUCGGGGGAGGAAUGGUAAAGCGAGCA----------AAA .(((((....)))))(((((((((((((((.((..(.(((.(.........).))).)...(((((((.....))))))).)).))))))...))))))))).----------... ( -37.40) >DroYak_CAF1 43063 106 - 1 GGCAGAACAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGGCGGAUGAGUUCGAGGAAAACUCGAGGGAGGAAUGGUACAGCGAGCA----------AAA .(((((....)))))((((((((..(((((.((..(.(((.(.........).))).)........(((((.....))))))).)))))..)...))))))).----------... ( -37.20) >DroAna_CAF1 31810 111 - 1 GGCAAAAGAUUUUGCGCUUGCUUUGUUUCUGCCGGGGCAUGGAAAAUUUUCGACUGGUGG-----UAGGAGGAGAAUCUGAUGGAAUCGAGGUUAAAAGGGAUUUGGGAAUUUAUC ......(((((((.(.((((((........(((((....((((.....)))).)))))))-----)))).).)))))))((((((.((.(((((......))))).))..)))))) ( -21.80) >consensus GGCAGAAUAUUUUGCGCUCGCUUUGUUUCUGCCAAGGCAUGCAAAAUUUUCGGAUGACGGUUGAGUUUGAGGAAAACUCGAGGGAGAAAUGGUAAAACGAGCA__________AAA .(((((....)))))(((((..(..(((((.((..(.(((.(.........).))).)...(((((((.....))))))).)).)))))..).....))))).............. (-20.79 = -22.35 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:58 2006