| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,040,575 – 13,040,677 |
| Length | 102 |
| Max. P | 0.682192 |

| Location | 13,040,575 – 13,040,677 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
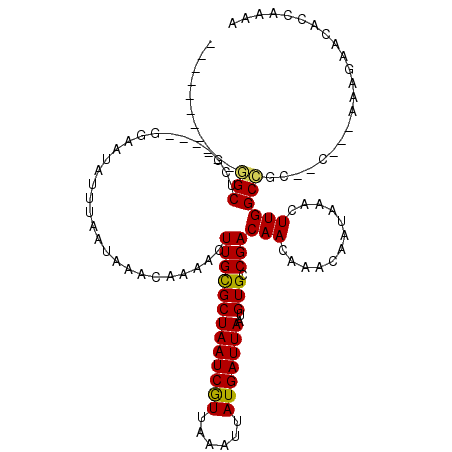
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -9.41 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
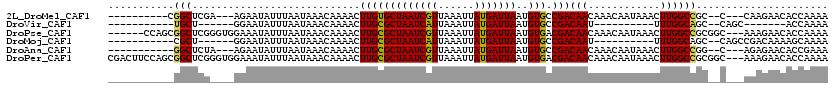
>2L_DroMel_CAF1 13040575 102 - 22407834 ----------CGGCUCGA---AGAAUAUUUAAUAAACAAAACUUGUGCUAAUCGUUAAAUUAUGAUUAAUGUGCCGACAACAAACAAUAAACUUGGCCGC--C---CAAGAACACCAAAA ----------((((.((.---..............(((.....)))..(((((((......))))))).)).))))...............(((((....--)---)))).......... ( -16.80) >DroVir_CAF1 23276 84 - 1 -----------UGCU------GGAAUAUUUAAUAAACAAAACUUGCGCUAAUCAUUAAAUUAUGAUUAAUGUGCCGACAAU----------UUUGGCAGC--CAGC-------ACCAAAA -----------((((------((.....................(((.(((((((......))))))).)))(((((....----------.)))))..)--))))-------)...... ( -22.30) >DroPse_CAF1 27905 111 - 1 ------CCAGCGGCUCGGGUGGAAAUAUUUAAUAAACAAAACUUGCGCUAAUCGUUAAAUUAUGAUUAAUGUGACGACAACAAACAAUAAACUUGGCCGCGGC---AAAGAACACCAAAA ------((.((((((.(((((.((..................)).)))...((((((.((((....)))).))))))..............)).)))))))).---.............. ( -21.57) >DroMoj_CAF1 27181 91 - 1 -----------CGCU------GGAAUAUUUAAUAAACAAAACUUGCGCUAAUCAUUAAAUUAUGAUUAAUGUGCCGACAAU----------UUUGGCAGC--CAGCCGACAAAAGCAAAA -----------.(((------((.....................(((.(((((((......))))))).)))(((((....----------.)))))..)--)))).............. ( -21.30) >DroAna_CAF1 14343 101 - 1 -----------GGCUCUA---AGAAUAUUUAAUAAACAAAACUUGCGCUAAUCGUUAAAUUAUGAUUAAUGUGCCGACAACAAACAAUAAACUUGGCCGG--C---AGAGAACACCGAAA -----------((((((.---.......................(((.(((((((......))))))).)))((((.(((............)))..)))--)---))))....)).... ( -17.10) >DroPer_CAF1 25103 117 - 1 CGACUUCCAGCGGCUCGGGUGGAAAUAUUUAAUAAACAAAACUUGCGCUAAUCGUUAAAUUAUGAUUAAUGUGACGACAACAAACAAUAAACUUGGCCGCGGC---AAAGAACACCAAAA ...(((((.((((((.(((((.((..................)).)))...((((((.((((....)))).))))))..............)).)))))))).---.))).......... ( -22.47) >consensus ___________GGCUCG____GGAAUAUUUAAUAAACAAAACUUGCGCUAAUCGUUAAAUUAUGAUUAAUGUGCCGACAACAAACAAUAAACUUGGCCGC__C___AAAGAACACCAAAA ...........(((............................(((((((((((((......)))))))..))).)))(((............))))))...................... ( -9.41 = -9.00 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:50 2006