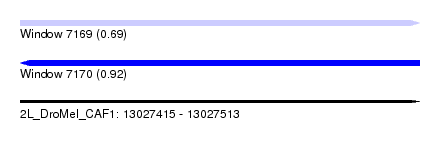
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,027,415 – 13,027,513 |
| Length | 98 |
| Max. P | 0.920984 |

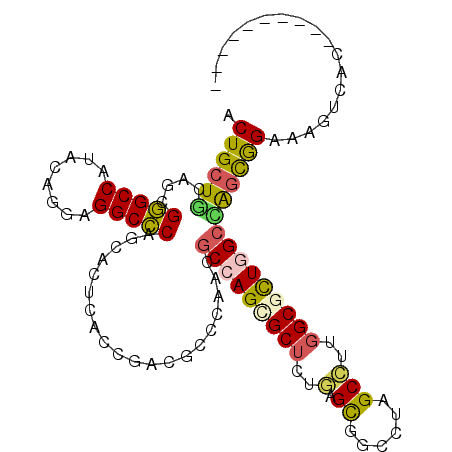
| Location | 13,027,415 – 13,027,513 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -46.68 |
| Consensus MFE | -28.62 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13027415 98 + 22407834 ----------GUGGCUUUCCGCCCGCCAACGCCAAAGCAAGUCCGCUUAGAGCGCUGGCGUUCGGAGUUGGCGAGUGCUGGGCCUCCUGUAUGGCCCGCUGCAGCAGA ----------(..((....(((.(((((((.((.((((......)))).(((((....))))))).))))))).)))..(((((........)))))))..)...... ( -46.80) >DroVir_CAF1 8745 108 + 1 GUGCCCGGCGGACACUUGCCGGUGGCUAGCGCCAAGGCCAACCCGCUCAAUGCACUGGCAUUGGGCGUCGGCGAAUGCUGUGCCUCCUGUAUGGCACGCUGCAGCAGA .(((.((((((.((.((((((((((((........)))).)))((((((((((....))))))))))..))))).))))(((((........)))))))))..))).. ( -50.90) >DroPse_CAF1 8155 98 + 1 ----------CUGACUUUCCGCUAGCCAGGGCCAAGGCUAGGCCACUCAGAGCGCUGGCGUUCGGUGUCGGUGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGU ----------..........(((.((..((((((...(((((...(((((..((((((((.....))))))))....)))))...))))).))))))...)))))... ( -42.20) >DroWil_CAF1 8324 98 + 1 ----------ACGACUUGCUACCGGCUAACGCCAGGGCCAGGCCACUAAGAGCACUGGCAUUGGGCGUUGGCGAUUGUUGGGCCUCCUGUAUGGCCCGUUGCAGCAGG ----------....((((((....(((((((((...(((((((........)).)))))....)))))))))....((((((((........))))))..)))))))) ( -47.30) >DroAna_CAF1 971 108 + 1 GCACCCGGCGGAUGUUUUCCGCCCGCCAACGCCAAGGCAAGGCCGCUCAGAGCGCUGGAGUUGGGCGUGGGUGAGUGUUGGGCCUCCUGUAUGGCCCGCUGAAGCAGU .(((((((((((.....))))))(((((((.(((.(((...)))((.....))..))).))).)))).)))))......(((((........)))))((....))... ( -50.70) >DroPer_CAF1 8162 98 + 1 ----------CUGACUUUCCGCUAGCCAGGGCCAAGGCUAGGCCACUCAGAGCGCUGGCGUUCGGUGUCGGUGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGU ----------..........(((.((..((((((...(((((...(((((..((((((((.....))))))))....)))))...))))).))))))...)))))... ( -42.20) >consensus __________GUGACUUUCCGCCAGCCAACGCCAAGGCAAGGCCACUCAGAGCGCUGGCGUUCGGCGUCGGCGAGUGCUGGGCCUCCUGUAUGGCCCGCUGCAGCAGU ........................(((.(((((((.(((((((........)).))))).)).))))).)))...(((((((((........))))).....)))).. (-28.62 = -28.48 + -0.14)

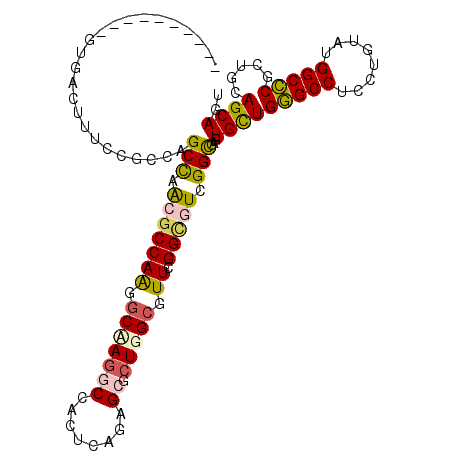
| Location | 13,027,415 – 13,027,513 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -28.57 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13027415 98 - 22407834 UCUGCUGCAGCGGGCCAUACAGGAGGCCCAGCACUCGCCAACUCCGAACGCCAGCGCUCUAAGCGGACUUGCUUUGGCGUUGGCGGGCGGAAAGCCAC---------- ...(((...(((((((........))))).))..........((((..((((((((((..(((((....))))).))))))))))..)))).)))...---------- ( -47.30) >DroVir_CAF1 8745 108 - 1 UCUGCUGCAGCGUGCCAUACAGGAGGCACAGCAUUCGCCGACGCCCAAUGCCAGUGCAUUGAGCGGGUUGGCCUUGGCGCUAGCCACCGGCAAGUGUCCGCCGGGCAC ..((((.(.(((((((........))))).(((((.((((.(((.((((((....)))))).)))(((((((......)))))))..)))).)))))..)).))))). ( -48.20) >DroPse_CAF1 8155 98 - 1 ACUGCUGCAGCGGGCCAUCCAGGAGGCCCAGCACUCACCGACACCGAACGCCAGCGCUCUGAGUGGCCUAGCCUUGGCCCUGGCUAGCGGAAAGUCAG---------- .(((((((...((((((...((((((((....(((((..(....)((.((....)).))))))))))))..)))))))))..)).)))))........---------- ( -36.60) >DroWil_CAF1 8324 98 - 1 CCUGCUGCAACGGGCCAUACAGGAGGCCCAACAAUCGCCAACGCCCAAUGCCAGUGCUCUUAGUGGCCUGGCCCUGGCGUUAGCCGGUAGCAAGUCGU---------- ..((((((...(((((........))))).......((.((((((....(((((.(((......))))))))...)))))).))..))))))......---------- ( -43.20) >DroAna_CAF1 971 108 - 1 ACUGCUUCAGCGGGCCAUACAGGAGGCCCAACACUCACCCACGCCCAACUCCAGCGCUCUGAGCGGCCUUGCCUUGGCGUUGGCGGGCGGAAAACAUCCGCCGGGUGC ...((....))(((((........)))))......(((((.((((.(((.((((.((.....))(((...))))))).)))))))((((((.....))))))))))). ( -50.20) >DroPer_CAF1 8162 98 - 1 ACUGCUGCAGCGGGCCAUCCAGGAGGCCCAGCACUCACCGACACCGAACGCCAGCGCUCUGAGUGGCCUAGCCUUGGCCCUGGCUAGCGGAAAGUCAG---------- .(((((((...((((((...((((((((....(((((..(....)((.((....)).))))))))))))..)))))))))..)).)))))........---------- ( -36.60) >consensus ACUGCUGCAGCGGGCCAUACAGGAGGCCCAGCACUCACCGACGCCCAACGCCAGCGCUCUGAGCGGCCUAGCCUUGGCGCUGGCCAGCGGAAAGUCAC__________ .((((((....(((((........)))))....................(((((((((..(.((......)))..))))))))))))))).................. (-28.57 = -28.10 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:48 2006