
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,025,129 – 13,025,301 |
| Length | 172 |
| Max. P | 0.945982 |

| Location | 13,025,129 – 13,025,248 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -52.92 |
| Consensus MFE | -29.31 |
| Energy contribution | -30.37 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13025129 119 - 22407834 CUGGAGCAGAUCUUCCACACACCGCUCUCCGCCAGCGCCGCAGCCGUGGUGAAUGCGGCCAACACCCAGCAGGAUAUCCAAGCGGCGGCAGCCUAUUGGGCAGCAGCCUGCAAAGCAGU- .(((((......)))))......(((..((((...((((((....))))))...))))..........(((((.(..((((..(((....)))..))))..)....)))))..)))...- ( -41.20) >DroVir_CAF1 6225 120 - 1 CUGGAGCAACUGCUGCAUUCACCGCUCUCCGCCAGCGCUGCGGCCGUGGUGAAUGCGGCCAAUACGCAGCAGGAUCUGCAAGCGGCAGCCGCCUAUUGGGCCGCUGCCUGCAAGGUGGUC ...((.((.((((((((((((((((...((((.......))))..))))))))))))))......(((((((...))))....((((((.(((.....))).))))))))).)).)).)) ( -58.60) >DroPse_CAF1 5899 119 - 1 UUGGAGCAGCUGCUGCACUCGCCGCUCUCUGCAGGCGCCGCGGCGGUGGUGAAUGCGGCCAACACACAGCAGGAUCUCCAGGCGGUGGCCGCCUACUGGGCAGCCGCCUGCAAGGCAGC- ((((((...((((((((.(((((((.(.((((.(....))))).)))))))).)))(.....)....)))))...))))))((((((((.(((.....))).)))))).))........- ( -54.60) >DroGri_CAF1 6105 120 - 1 CUGGAGCAAUUGUUGCAUUCACCGCUCUCUGCCAGUGCCGCUGCUGUGGUGAAUGCCGCAAAUACGCAACAGGAUUUGCAGGCAGCUGCUGCAUAUUGGGCCGCUGCCUGCAAAGCAGCA (((..((..((((.(((((((((((.....((.((.....)))).))))))))))).))))....))..)))..((((((((((((.(((........))).))))))))))))...... ( -55.30) >DroWil_CAF1 5252 119 - 1 UUGGAGCAACUACUUCAUUCCCCACUCUCGGCCAGUGCUGCGGCUGUGGUAAAUGGUGCCAAUACCUCACAGGAUCUGCAAGCAGUGGCCGCCAUUUGGGCAGCAGCCUGCAAGGCAGC- .(((((......)))))...((((....((((((.((((((((((((((.....((((....))))))))))...)))).)))).)))))).....))))..((.(((.....))).))- ( -45.10) >DroMoj_CAF1 5942 120 - 1 CUGGAGCAGCUGCUCCACUCGCCCCUCUCCGCCAGCACCGCAGCCGUGGUGAAUGCAGCCAACACGCAGCAGGAUCUGGAGGUGGCGGCCGCCUGCUGGGCCGCUGCCUAUAAGGCGGCC .((((((....))))))...((((((((...((..((((((....))))))..(((.((......)).)))))....))))).)))((((((((..(((((....)))))..)))))))) ( -62.70) >consensus CUGGAGCAACUGCUGCACUCACCGCUCUCCGCCAGCGCCGCGGCCGUGGUGAAUGCGGCCAACACGCAGCAGGAUCUGCAAGCGGCGGCCGCCUAUUGGGCAGCUGCCUGCAAGGCAGC_ .....((....((((((.(((((((...(.((.......)).)..))))))).)))))).................((((.(((((.(((........))).))))).))))..)).... (-29.31 = -30.37 + 1.06)

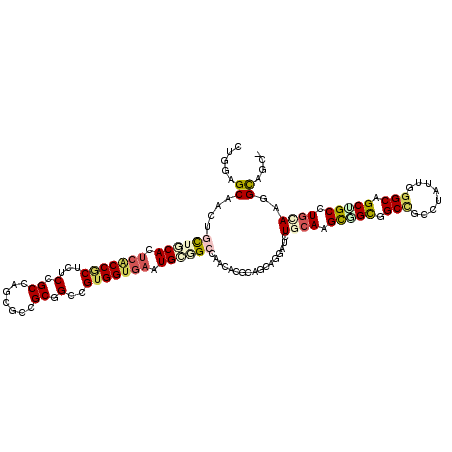
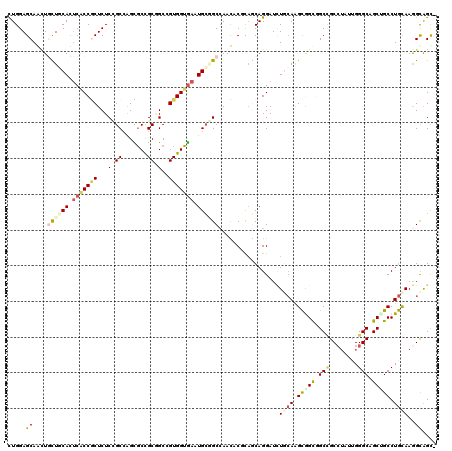
| Location | 13,025,168 – 13,025,279 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13025168 111 - 22407834 ---------UGCGCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACACCGCUCUCCGCCAGCGCCGCAGCCGUGGUGAAUGCGGCCAACACCCAGCAGGAUAUCCA ---------(((((.((.((.((((((........)))))).)).)).(......).......))...((((...((((((....))))))...))))..........)))((....)). ( -37.60) >DroVir_CAF1 6265 111 - 1 CCUGAAUGGCGCC---------UUGCUUAGCAGCAAACAGCUGGAGCAACUGCUGCAUUCACCGCUCUCCGCCAGCGCUGCGGCCGUGGUGAAUGCGGCCAAUACGCAGCAGGAUCUGCA ((((....(((..---------((((((..((((.....))))))))))..((((((((((((((...((((.......))))..)))))))))))))).....)))..))))....... ( -50.80) >DroPse_CAF1 5938 111 - 1 ---------UGCCCAACAGCUGCUGAUGAGCUCCAAGCAAUUGGAGCAGCUGCUGCACUCGCCGCUCUCUGCAGGCGCCGCGGCGGUGGUGAAUGCGGCCAACACACAGCAGGAUCUCCA ---------..........((((((.((.(((((((....)))))))....((((((.(((((((.(.((((.(....))))).)))))))).)))))).....))))))))........ ( -51.20) >DroGri_CAF1 6145 102 - 1 ---------UUCA---------GUGCUGAGCAGCAAGCAACUGGAGCAAUUGUUGCAUUCACCGCUCUCUGCCAGUGCCGCUGCUGUGGUGAAUGCCGCAAAUACGCAACAGGAUUUGCA ---------((((---------(((((........))).))))))....((((.(((((((((((.....((.((.....)))).))))))))))).))))....((((......)))). ( -36.50) >DroMoj_CAF1 5982 102 - 1 ---------CGGC---------CUGCUGAGCAGCAAGCAGCUGGAGCAGCUGCUCCACUCGCCCCUCUCCGCCAGCACCGCAGCCGUGGUGAAUGCAGCCAACACGCAGCAGGAUCUGGA ---------((((---------((((((....((..((((.((((((....)))))))).))........((...((((((....))))))...)).)).......)))))))..))).. ( -39.40) >DroPer_CAF1 5951 111 - 1 ---------UGCCCAACAGCUGCUGAUGAGCUCCAAGCAAUUGGAGCAGCUGCUGCACUCGCCCCUCUCUGCAGGCGCCGCGGCGGUGGUGAAUGCGGCCAACACACAGCAGGAUCUCCA ---------..........((((((.((.(((((((....)))))))....((((((.(((((((...((((.(....))))).)).))))).)))))).....))))))))........ ( -47.90) >consensus _________UGCC_________CUGCUGAGCACCAAGCAACUGGAGCAGCUGCUGCACUCACCGCUCUCCGCCAGCGCCGCGGCCGUGGUGAAUGCGGCCAACACGCAGCAGGAUCUCCA ......................(((.((..............(((((.((..........)).)))))((((...((((((....))))))...))))........)).)))........ (-17.00 = -17.87 + 0.86)

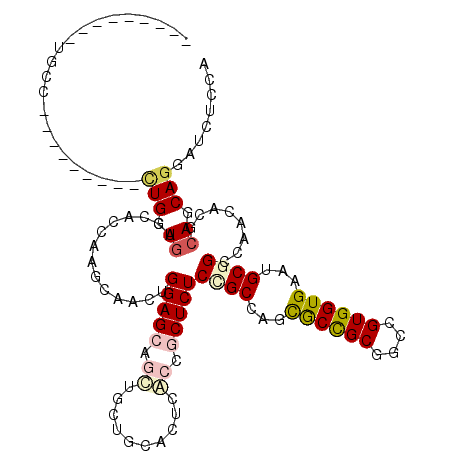
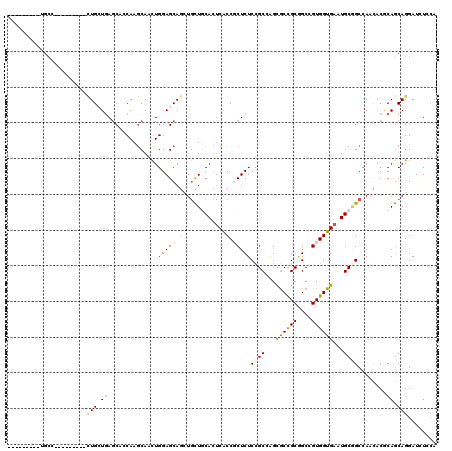
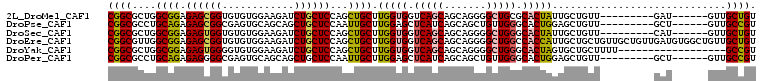
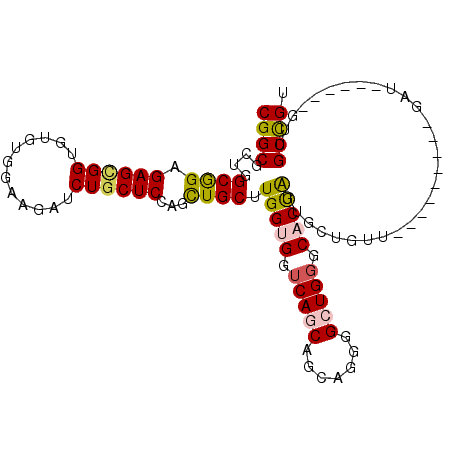
| Location | 13,025,208 – 13,025,301 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -42.63 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13025208 93 + 22407834 CGGCGCUGGCGGAGAGCGGUGUGUGGAAGAUCUGCUCCAGCUGCUUGGUGGUCAGCAGCAGGGGCUGCGCACUAUUGCUGUU---------GAU------GUUGCUGU ((((((((((.(.(((((((..(..(.....)..)....)))))))..).))))))((((..(((.(((......))).)))---------..)------))))))). ( -36.10) >DroPse_CAF1 5978 93 + 1 CGGCGCCUGCAGAGAGCGGCGAGUGCAGCAGCUGCUCCAAUUGCUUGGAGCUCAUCAGCAGCUGUUGGGCACUGGAGCUGUU---------GCU------GUUGCCGU (((((((.((.....)))))....((((((((.((((((..((((..((((((....).)))).)..)))).)))))).)))---------)))------)).)))). ( -47.80) >DroSec_CAF1 6743 93 + 1 CGGCGCUGGCGGAGAGUGGUGUGUGGAAGAUCUGCUCCAGCUGCUUGGUGGUCAGCAGCAGGGGCUGGGCACUAUUGCUGUU---------CAU------GUUGCUGU ((((((((((((..(((((((((..(.....)..).((((((.((((.((.....)).)))))))))))))))))).))).)---------)).------).))))). ( -35.90) >DroEre_CAF1 6789 108 + 1 CGGCGUUGGCGGAGAGCGGUGUGUGGAAGAUCUGCUCCAGCUGCUUGGUGGUCAGCAGCAGGGGCUGGCCACCAUUGCUGCUGUUGCUGUUGAUGUGGCUGUUGCUGU ..((((..((((.((((((............))))))((((.((.(((((((((((.......)))))))))))..)).))))...))))..))))(((....))).. ( -50.60) >DroYak_CAF1 5487 90 + 1 CGGCGCUGGCGGAGAGUGGGGUGUGGAAGAUCUGCUCCAGCUGCUUGGUGGUCAGCAGCAGGGGCUGGGCACUAGUGCUGCUUUU------------------GCCGU (((((..(((((..((((((((.......))))(((((.((((((........)))))).)))))....))))....)))))..)------------------)))). ( -38.70) >DroPer_CAF1 5991 93 + 1 CGGCGCCUGCAGAGAGGGGCGAGUGCAGCAGCUGCUCCAAUUGCUUGGAGCUCAUCAGCAGCUGUUGGGCACUGGAGCUGUU---------GCU------GUUGCCGU ((((((((........))))....((((((((.((((((..((((..((((((....).)))).)..)))).)))))).)))---------)))------)).)))). ( -46.70) >consensus CGGCGCUGGCGGAGAGCGGUGUGUGGAAGAUCUGCUCCAGCUGCUUGGUGGUCAGCAGCAGGGGCUGGGCACUAGUGCUGUU_________GAU______GUUGCCGU ((((....((((.((((((............))))))...)))).(((((.(((((.......))))).))))).............................)))). (-24.90 = -24.68 + -0.22)
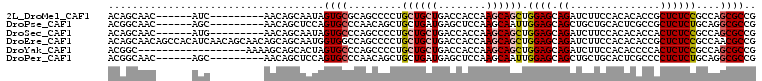
| Location | 13,025,208 – 13,025,301 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.64 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 13025208 93 - 22407834 ACAGCAAC------AUC---------AACAGCAAUAGUGCGCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACACCGCUCUCCGCCAGCGCCG ...((...------...---------....))....(.((((.((.((.((((((........)))))).)).))................((.....))..))))). ( -25.30) >DroPse_CAF1 5978 93 - 1 ACGGCAAC------AGC---------AACAGCUCCAGUGCCCAACAGCUGCUGAUGAGCUCCAAGCAAUUGGAGCAGCUGCUGCACUCGCCGCUCUCUGCAGGCGCCG .((((...------.((---------(.(((((.((((((......)).))))....(((((((....)))))))))))).)))....(((((.....)).))))))) ( -43.20) >DroSec_CAF1 6743 93 - 1 ACAGCAAC------AUG---------AACAGCAAUAGUGCCCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACACCACUCUCCGCCAGCGCCG ...((...------...---------....))....((((...((....((((((........)))))).((((.((...............))))))))..)))).. ( -20.66) >DroEre_CAF1 6789 108 - 1 ACAGCAACAGCCACAUCAACAGCAACAGCAGCAAUGGUGGCCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACACCGCUCUCCGCCAACGCCG ...((................((....)).....((((((..(((....((((((........))))))(((((......)))))......)))..))))))..)).. ( -29.70) >DroYak_CAF1 5487 90 - 1 ACGGC------------------AAAAGCAGCACUAGUGCCCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACCCCACUCUCCGCCAGCGCCG .((((------------------...(((((((...((.....))...)))))))............(((((.(.(((...............))).).))))))))) ( -27.36) >DroPer_CAF1 5991 93 - 1 ACGGCAAC------AGC---------AACAGCUCCAGUGCCCAACAGCUGCUGAUGAGCUCCAAGCAAUUGGAGCAGCUGCUGCACUCGCCCCUCUCUGCAGGCGCCG .((((...------.((---------(.(((((.((((((......)).))))....(((((((....)))))))))))).)))....(((..........))))))) ( -39.30) >consensus ACAGCAAC______AUC_________AACAGCAAUAGUGCCCAGCCCCUGCUGCUGACCACCAAGCAGCUGGAGCAGAUCUUCCACACACCGCUCUCCGCCAGCGCCG ....................................((((.........((((((........)))))).((((.((...............))))))....)))).. (-13.25 = -13.64 + 0.39)
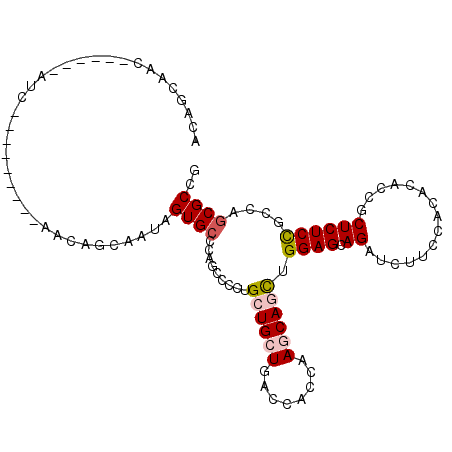
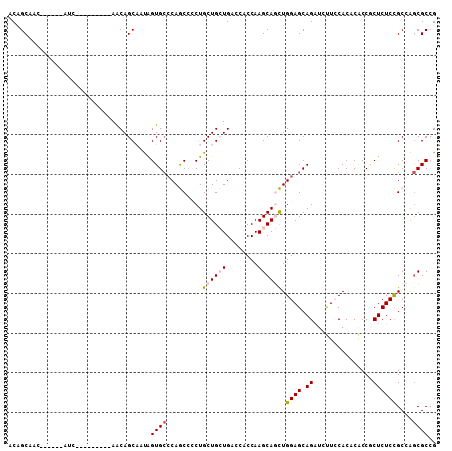
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:42 2006