| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 13,023,109 – 13,023,201 |
| Length | 92 |
| Max. P | 0.901801 |

| Location | 13,023,109 – 13,023,201 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -21.07 |
| Consensus MFE | -7.93 |
| Energy contribution | -10.04 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
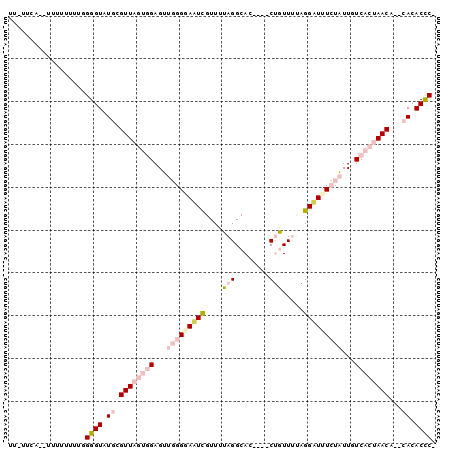
>2L_DroMel_CAF1 13023109 92 + 22407834 -GGGUGUG--UGUUAGUGACAAUAGAAAUCCCAAAACAG----GUGCCUAAAACGAAUCCCCAACUCCACUAACGCAUACCCAAAAAAAAAUAUGGA-AA -(((((((--((((((((....(((..(((........)----))..)))....(......).....)))))))))))))))...............-.. ( -26.10) >DroPse_CAF1 3875 92 + 1 GGAGUGUG--UGUUAGUGACAAAAUAUAACCAUUAAUCGGUGUGUGCCUAAAACAAUUCCCAACCUC----AACAGAAACCCGAAAAAAAA--UAAACUA (((((.((--(.((((..(((.......(((.......))).)))..)))).)))))))).......----....................--....... ( -12.60) >DroSec_CAF1 4646 90 + 1 -GGGUGUG--UGUUAGUGACAAUAGAAAUCCUAAAACAG----GUGCCUAAAACGAUUUCCCAACUCCACUAACGCAUACCCCAAAAAAAA--UGGA-AA -(((((((--((((((((......((((((.......((----....)).....)))))).......))))))))))))))).........--....-.. ( -28.22) >DroEre_CAF1 4697 89 + 1 -GGGUGUG--UGUUAGUGACAAUAGAAAUCCUAAAACAG----GUGCCUAAAACGAAUCCCCAACUCCACUAACGCAUACCCCAAAAAAAA--GGAA--A -(((((((--((((((((....(((....(((.....))----)...)))....(......).....))))))))))))))).........--....--. ( -25.80) >DroYak_CAF1 3405 92 + 1 -GGGUGUGUUUGUUAGUGACAAUAGAAAUCCUAAAACAG----GUGCCUAAAACGAAUCCCCAACUCCACUAACGCAUACCCCAAAAAAAA--GGAA-AA -((((((((..(((((((....(((....(((.....))----)...)))....(......).....))))))))))))))).........--....-.. ( -21.10) >DroPer_CAF1 3891 91 + 1 GGAGUGUG--UGUUAGUGACAAAAUAUAACCAUUAAUCGGUGUGUGCCUAAAACAAUUCCCAACCUC----AACAGAAACCCGAAA-AAAA--UAAACUA (((((.((--(.((((..(((.......(((.......))).)))..)))).)))))))).......----...............-....--....... ( -12.60) >consensus _GGGUGUG__UGUUAGUGACAAUAGAAAUCCUAAAACAG____GUGCCUAAAACGAAUCCCCAACUCCACUAACGCAUACCCCAAAAAAAA__UGAA_AA .(((((((..((((((((.................................................))))))))))))))).................. ( -7.93 = -10.04 + 2.11)


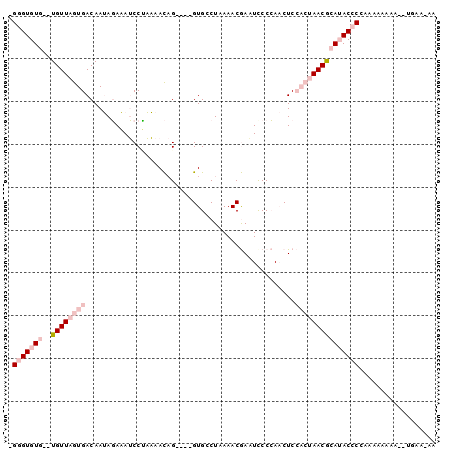
| Location | 13,023,109 – 13,023,201 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.15 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -12.56 |
| Energy contribution | -14.98 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
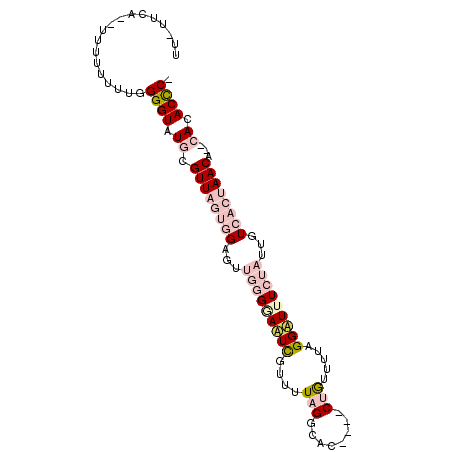
>2L_DroMel_CAF1 13023109 92 - 22407834 UU-UCCAUAUUUUUUUUUGGGUAUGCGUUAGUGGAGUUGGGGAUUCGUUUUAGGCAC----CUGUUUUGGGAUUUCUAUUGUCACUAACA--CACACCC- ..-...............((((.((.((((((((...(((..((((....(((....----))).....))))..)))...)))))))).--)).))))- ( -28.30) >DroPse_CAF1 3875 92 - 1 UAGUUUA--UUUUUUUUCGGGUUUCUGUU----GAGGUUGGGAAUUGUUUUAGGCACACACCGAUUAAUGGUUAUAUUUUGUCACUAACA--CACACUCC ..(((((--..(..((((.((..((....----))..)).))))..)...))))).......(((.((((....))))..))).......--........ ( -10.20) >DroSec_CAF1 4646 90 - 1 UU-UCCA--UUUUUUUUGGGGUAUGCGUUAGUGGAGUUGGGAAAUCGUUUUAGGCAC----CUGUUUUAGGAUUUCUAUUGUCACUAACA--CACACCC- ..-....--.........((((.((.((((((((.....(((((((.(..(((....----)))....).)))))))....)))))))).--)).))))- ( -28.00) >DroEre_CAF1 4697 89 - 1 U--UUCC--UUUUUUUUGGGGUAUGCGUUAGUGGAGUUGGGGAUUCGUUUUAGGCAC----CUGUUUUAGGAUUUCUAUUGUCACUAACA--CACACCC- .--....--.........((((.((.((((((((...(((..((((....(((....----))).....))))..)))...)))))))).--)).))))- ( -29.40) >DroYak_CAF1 3405 92 - 1 UU-UUCC--UUUUUUUUGGGGUAUGCGUUAGUGGAGUUGGGGAUUCGUUUUAGGCAC----CUGUUUUAGGAUUUCUAUUGUCACUAACAAACACACCC- ..-....--.........((((.((.((((((((...(((..((((....(((....----))).....))))..)))...))))))))...)).))))- ( -26.30) >DroPer_CAF1 3891 91 - 1 UAGUUUA--UUUU-UUUCGGGUUUCUGUU----GAGGUUGGGAAUUGUUUUAGGCACACACCGAUUAAUGGUUAUAUUUUGUCACUAACA--CACACUCC .......--....-((((.((..((....----))..)).)))).((((...((((...((((.....)))).......))))...))))--........ ( -10.10) >consensus UU_UUCA__UUUUUUUUGGGGUAUGCGUUAGUGGAGUUGGGGAAUCGUUUUAGGCAC____CUGUUUUAGGAUUUCUAUUGUCACUAACA__CACACCC_ ..................((((.((.((((((((...(((((((((....(((........))).....)))))))))...))))))))...)).)))). (-12.56 = -14.98 + 2.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:37 2006