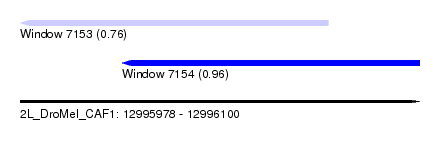
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,995,978 – 12,996,100 |
| Length | 122 |
| Max. P | 0.960056 |

| Location | 12,995,978 – 12,996,072 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
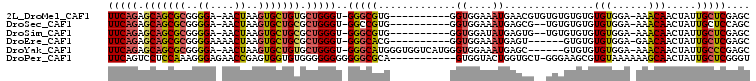
>2L_DroMel_CAF1 12995978 94 - 22407834 UUCAGAGCAGCGCGGGGA-AACUAAGUGCUGUGCUGGGU-GGGCGUG----------GGUGGAAAUGAACGUGUGUGUGUGUGUGGA-AAACAACUAUUGCUCGAGC (((((.(((((((.((..-..))..))))))).)))))(-(((((((----------(.((.......(((..(....)..)))...-...)).))).))))))... ( -29.62) >DroSec_CAF1 21215 92 - 1 UUCAGAGCAGCGCGGGGA-AACUAAGUGCUGCGCUGGGU-GGCCGUG----------GGUGGAAAUGAGCG--UGUGUGUGUGUGGA-AAACAACUAUUGCUCCAGC ......(((((((.((..-..))..)))))))(((((..-(((.(((----------(.((.......(((--......))).....-...)).)))).)))))))) ( -30.96) >DroSim_CAF1 19968 92 - 1 UUCAGAGCAGCGCGGGGA-AACUAAGUGCUGCGCUGGGU-GGGCGUG----------GGUGGAUAUGAGUG--UGUGUGUGUGUGGA-AAACAACUAUUGCUCGAGC (((((.(((((((.((..-..))..))))))).)))))(-(((((((----------(..(.((((.....--.)))).).(((...-..))).))).))))))... ( -29.40) >DroEre_CAF1 22828 89 - 1 UUCAGAGCAGCGCGGGGAAAACUAAGUGCUGCGCUGGGU-GGGCACG----------GGUGGAAAUGAGU------GUGUGUGUGGA-GAACAACUAUUGCUCGAGC ......(((((((..((....))..)))))))((((((.-.(((((.----------.((....))..))------))...(((...-..)))....)..))).))) ( -28.30) >DroYak_CAF1 20253 98 - 1 UUCAGAGCAGCGCGGGGA-AACUAAGUGCUGUGCUGGGU-GGGCAUGGGUGGUCAUGGGUGGAAAUGAGC------GUGUGUGUGGA-AAACAACUAUUGCCCGAGC (((((.(((((((.((..-..))..))))))).)))))(-(((((..((((.((((........)))).)------.....(((...-..))))))..))))))... ( -35.10) >DroPer_CAF1 41127 95 - 1 UUCAGUCCUCCAAAGGGAGAACCGAGUGGUGUGGGGGGGGGGGCGCA-----------GUGGUACUGGUGCU-GGGAAGCGUGUAAAAAAGCAACUAUUGCUCGGGU ....(((((((.........(((....))).......)))))))(((-----------(((((......(((-....))).(((......)))))))))))...... ( -27.99) >consensus UUCAGAGCAGCGCGGGGA_AACUAAGUGCUGCGCUGGGU_GGGCGUG__________GGUGGAAAUGAGCG__UGUGUGUGUGUGGA_AAACAACUAUUGCUCGAGC (((((.(((((((..((....))..))))))).)))))..(((((.............((....))...............(((......))).....))))).... (-19.53 = -19.48 + -0.05)
| Location | 12,996,009 – 12,996,100 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -26.27 |
| Energy contribution | -27.58 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
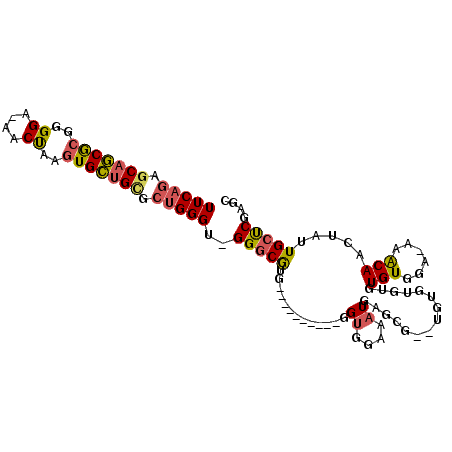
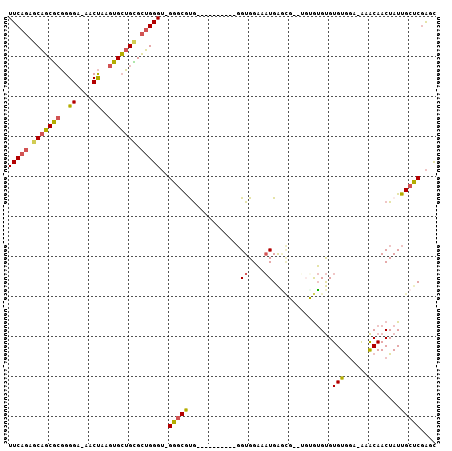
>2L_DroMel_CAF1 12996009 91 - 22407834 --------ACCCACCACCCCGUUGGCACGUCAUAAUUUCAGAGCAGCGCGGGGA-AACUAAGUGCUGUGCUGGGU-GGGCGUG----------GGUGGAAAUGAACGUGUG --------..(((((.((.....))((((((.....(((((.(((((((.((..-..))..))))))).))))).-.))))))----------)))))............. ( -36.90) >DroSec_CAF1 21246 95 - 1 --GCCACCACCCACCAUCCCGUUGGCACGUCAUAAUUUCAGAGCAGCGCGGGGA-AACUAAGUGCUGCGCUGGGU-GGCCGUG----------GGUGGAAAUGAGCG--UG --((((((((((((((......))....(((((....((((.(((((((.((..-..))..))))))).))))))-))).)))----------)))))...)).)).--.. ( -41.50) >DroSim_CAF1 19999 95 - 1 --CCCACCACCCACCACCCCGUUGGCACGUCAUAAUUUCAGAGCAGCGCGGGGA-AACUAAGUGCUGCGCUGGGU-GGGCGUG----------GGUGGAUAUGAGUG--UG --....((((((((..((((..(((....)))......(((.(((((((.((..-..))..))))))).))))).-))..)))----------))))).........--.. ( -42.40) >DroEre_CAF1 22858 95 - 1 CACCCAGCACCCACCACCCCGUUGGCACGUCAUAAUUUCAGAGCAGCGCGGGGAAAACUAAGUGCUGCGCUGGGU-GGGCACG----------GGUGGAAAUGAGU----- (((((.(..((((((....(((....))).........(((.(((((((..((....))..))))))).))))))-)))..))----------)))).........----- ( -39.30) >DroYak_CAF1 20283 102 - 1 --CCCAGCAGCCACCACCCCGUCGGCACGUCAUAAUUUCAGAGCAGCGCGGGGA-AACUAAGUGCUGUGCUGGGU-GGGCAUGGGUGGUCAUGGGUGGAAAUGAGC----- --((((...((((((..(((..(((....)).......(((.(((((((.((..-..))..))))))).))))..-)))....))))))..))))...........----- ( -41.60) >DroPer_CAF1 41159 98 - 1 UAUCCUGCU-CUCCCACCCCUUUGGCACGUCAUAAUUUCAGUCCUCCAAAGGGAGAACCGAGUGGUGUGGGGGGGGGGGCGCA-----------GUGGUACUGGUGCU-GG ((((((((.-(((((.(((((....(((..(((....((.((.((((....)))).)).)))))..))))))))))))).)))-----------).))))........-.. ( -41.20) >consensus __CCCAGCACCCACCACCCCGUUGGCACGUCAUAAUUUCAGAGCAGCGCGGGGA_AACUAAGUGCUGCGCUGGGU_GGGCGUG__________GGUGGAAAUGAGCG__UG ..........(((((..........((((((.....(((((.(((((((..((....))..))))))).)))))...))))))..........)))))............. (-26.27 = -27.58 + 1.31)

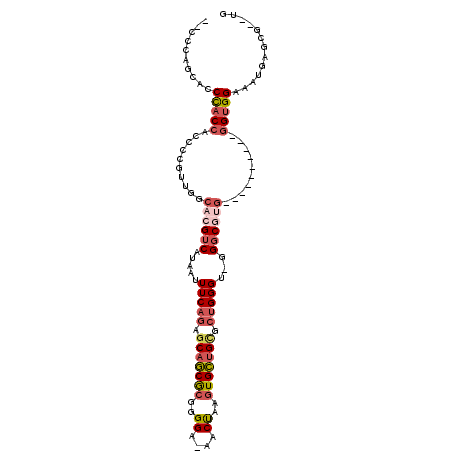
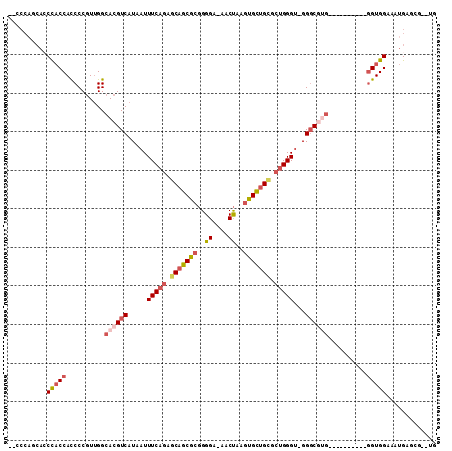
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:32 2006