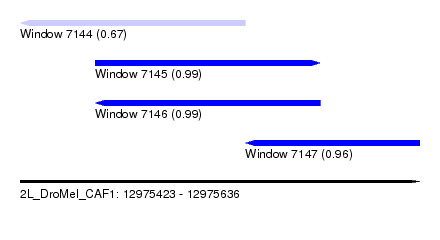
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,975,423 – 12,975,636 |
| Length | 213 |
| Max. P | 0.986032 |

| Location | 12,975,423 – 12,975,543 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.43 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975423 120 - 22407834 ACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUAAGUUUUUGUAGAGCCCACAGCUCCCAAAUUUUUCGAUUAU (((....((((...((.............((((..((((....))))..))))(((((((.....)))))))..))..))))....))).((((.....))))................. ( -27.20) >DroSec_CAF1 20055 119 - 1 ACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUAAGUUUUUGGAGAGUCCACAGCUCUCG-AUUUUUCGAUUAU ..............((((...((((..(.((((..((((....))))..))))(((((((.....)))))))...)..))))...)))).((((.....))))(((-(....)))).... ( -30.30) >DroSim_CAF1 21218 119 - 1 ACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUAAGUUUUUGGAGAGUCCACAGCUCUCG-AUUUUUCGAUUAU ..............((((...((((..(.((((..((((....))))..))))(((((((.....)))))))...)..))))...)))).((((.....))))(((-(....)))).... ( -30.30) >DroYak_CAF1 23041 108 - 1 CCACAUUCUCAUCACUAAUUUCUGGCUUGGCCCUCUGUCAGCUGACAAAGGGCAGGUGUAUCUGCUGCAUUUACAGGUUAAG-----------UCCACAGCUCCCU-AUUUUUCGAUUAU ......................((((((((((((.((((....)))).)))))(((((((.....))))))).)))))))..-----------.............-............. ( -24.70) >consensus ACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUAAGUUUUUGGAGAGUCCACAGCUCCCG_AUUUUUCGAUUAU ..........(((........((((....(((((.((((....)))).)))))(((((((.....))))))).))))...........((((((.....)))))).........)))... (-21.99 = -23.43 + 1.44)


| Location | 12,975,463 – 12,975,583 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -31.17 |
| Energy contribution | -30.80 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975463 120 + 22407834 UAACCUGUAAAUGUAGCAGAUACACCUGCCCCUUGUCAGCUGACAGAGGGCUGAGCAAGAAAUUAGUGAUAAGAAAAUGUUCACCUUUAUCUUCGCCCUUUUGCAGCCAGCAUUUAACAA .....(((((((((.((((......)))).........((((..(((((((((((((.....(((....))).....))))))...........)))))))..))))..)))))).))). ( -31.80) >DroSec_CAF1 20094 120 + 1 UAACCUGUAAAUGUAGCAGAUACACCUGCCCCUUGUCAGCUGACAGAGGGCUGAGCAAGAAAUUAGUGAUAAGAAAAUGUUCACCUUUAUCUUUGCCCUUUUCCAGCCAGCAUUUAACAA .....(((((((((.((((......)))).........((((..(((((((((((((.....(((....))).....))))))...........)))))))..))))..)))))).))). ( -31.70) >DroSim_CAF1 21257 120 + 1 UAACCUGUAAAUGUAGCAGAUACACCUGCCCCUUGUCAGCUGACAGAGGGCUGAGCAAGAAAUUAGUGAUAAGAAAAUGUUCACCUUUAUCUUCGCCCUUUUCCAGCCAGCAUUUAACAA .....(((((((((.((((......)))).........((((..(((((((((((((.....(((....))).....))))))...........)))))))..))))..)))))).))). ( -31.70) >DroYak_CAF1 23069 120 + 1 UAACCUGUAAAUGCAGCAGAUACACCUGCCCUUUGUCAGCUGACAGAGGGCCAAGCCAGAAAUUAGUGAUGAGAAUGUGGUCACCUUUAUCUUUGCCCUUUUGCAGCCAGCAUUUUACAA ....(((((((.((((.(((((.....((((((((((....))))))))))...((((.(..(((....)))...).))))......)))))))))...))))))).............. ( -36.20) >consensus UAACCUGUAAAUGUAGCAGAUACACCUGCCCCUUGUCAGCUGACAGAGGGCUGAGCAAGAAAUUAGUGAUAAGAAAAUGUUCACCUUUAUCUUCGCCCUUUUCCAGCCAGCAUUUAACAA .....(((((((((.((((......)))).........((((..(((((((.(((....(((...((((...........)))).)))..))).)))))))..))))..)))))).))). (-31.17 = -30.80 + -0.37)

| Location | 12,975,463 – 12,975,583 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -35.11 |
| Energy contribution | -34.80 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975463 120 - 22407834 UUGUUAAAUGCUGGCUGCAAAAGGGCGAAGAUAAAGGUGAACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUA ((((.(((((.((((((((...((((.((((....(((((...........)))))....)))))))).((((..((((....))))..))))...))))...))))))))))))).... ( -38.10) >DroSec_CAF1 20094 120 - 1 UUGUUAAAUGCUGGCUGGAAAAGGGCAAAGAUAAAGGUGAACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUA ((((.(((((.((((.(((...((((((.((....(((((...........)))))....)))))))).((((..((((....))))..)))).......)))))))))))))))).... ( -38.40) >DroSim_CAF1 21257 120 - 1 UUGUUAAAUGCUGGCUGGAAAAGGGCGAAGAUAAAGGUGAACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUA ((((.(((((.((((.(((...((((.((((....(((((...........)))))....)))))))).((((..((((....))))..)))).......)))))))))))))))).... ( -38.40) >DroYak_CAF1 23069 120 - 1 UUGUAAAAUGCUGGCUGCAAAAGGGCAAAGAUAAAGGUGACCACAUUCUCAUCACUAAUUUCUGGCUUGGCCCUCUGUCAGCUGACAAAGGGCAGGUGUAUCUGCUGCAUUUACAGGUUA (((((((.(((.((((((......))).(((((..(((((........)))))...........(((((.((((.((((....)))).))))))))).)))))))))))))))))).... ( -37.30) >consensus UUGUUAAAUGCUGGCUGCAAAAGGGCAAAGAUAAAGGUGAACAUUUUCUUAUCACUAAUUUCUUGCUCAGCCCUCUGUCAGCUGACAAGGGGCAGGUGUAUCUGCUACAUUUACAGGUUA ((((.(((((.((((.......((((.((((....(((((...........)))))....)))))))).(((((.((((....)))).)))))..........))))))))))))).... (-35.11 = -34.80 + -0.31)



| Location | 12,975,543 – 12,975,636 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -19.27 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975543 93 - 22407834 ---------------------------UUUGCUUUAUUCUGUUUUUUCGACUGCAAUGGAGGGAAAAUAGAAGAGGAAAAUUGUUAAAUGCUGGCUGCAAAAGGGCGAAGAUAAAGGUGA ---------------------------((((((((.(((((((((..(..(......)..)..)))))))))................(((.....)))..))))))))........... ( -21.80) >DroSim_CAF1 21337 120 - 1 AUCAUCGAAACGCUUAAGGUUGGACAUUUUGCUUUAUUCUGUUUUUUCGACUGCAAUGGAGGGAAAAUAGAAGAGGAAAAUUGUUAAAUGCUGGCUGGAAAAGGGCGAAGAUAAAGGUGA .(((((....(((((..(((..(.(((((.(((((.(((((((((..(..(......)..)..)))))))))...)))....)).))))))..)))......)))))........))))) ( -29.40) >DroYak_CAF1 23149 93 - 1 ---------------------------UUUGCUUUAUUCUGUUUUAACGACUGCAAUGGAGGGAAAAUAGAGGAGGAAAAUUGUAAAAUGCUGGCUGCAAAAGGGCAAAGAUAAAGGUGA ---------------------------((((((((.(((((((((..(..(......)..)..)))))))))................(((.....)))..))))))))........... ( -19.20) >consensus ___________________________UUUGCUUUAUUCUGUUUUUUCGACUGCAAUGGAGGGAAAAUAGAAGAGGAAAAUUGUUAAAUGCUGGCUGCAAAAGGGCGAAGAUAAAGGUGA ...........................((((((((.(((((((((..(..(......)..)..)))))))))................(((.....)))..))))))))........... (-19.27 = -19.17 + -0.11)

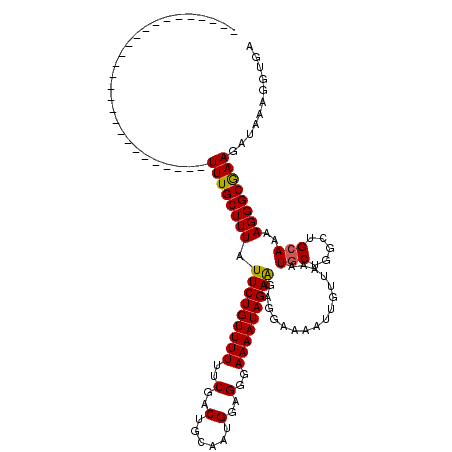
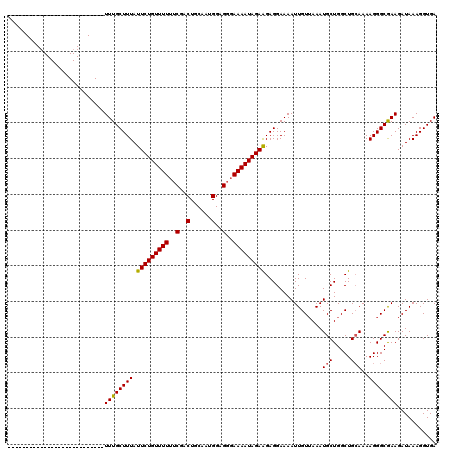
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:26 2006