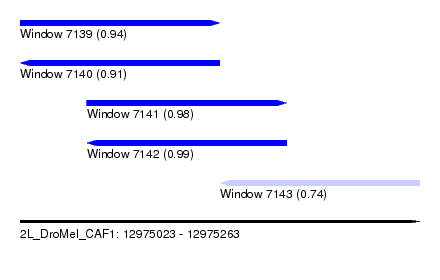
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,975,023 – 12,975,263 |
| Length | 240 |
| Max. P | 0.989684 |

| Location | 12,975,023 – 12,975,143 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -42.17 |
| Energy contribution | -42.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
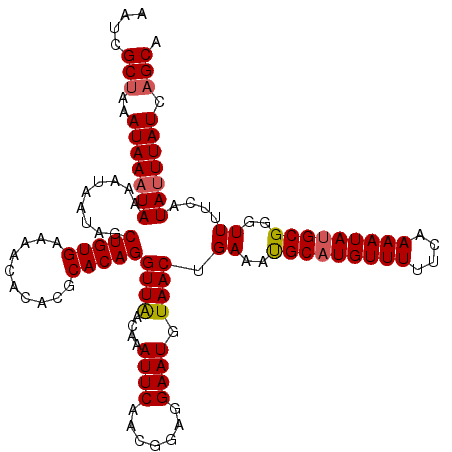
>2L_DroMel_CAF1 12975023 120 + 22407834 CAAUGACAGUAGUCUUGUGAUCAACGUCAAAAGCAAUUGUCAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUU ...(((((((.(.(((.((((....)))).)))).)))))))...(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).. ( -44.30) >DroSec_CAF1 19657 120 + 1 CAAUGACAGUAGUCUUGUGAUCAACGUCAAAAGCAAUUGUCAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUU ...(((((((.(.(((.((((....)))).)))).)))))))...(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).. ( -44.30) >DroSim_CAF1 20820 120 + 1 CAAUGACAGUAGUCUUGUGAUCAACGUCAAAAGCAAUUGUCAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUU ...(((((((.(.(((.((((....)))).)))).)))))))...(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).. ( -44.30) >DroYak_CAF1 22647 119 + 1 CAAUGACAGUAGUCUUGUGAUCAACGUCAAAAGCAAUUGUCAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCGUAGCGGAGAGUUACUCUCCCACUCCACUCUCUCUUGUUUUU-UU ...(((((((.(.(((.((((....)))).)))).)))))))..((((((....))))))(((((((((((((..(.((.((((((...))))))..)).)...)))))))))))))-.. ( -39.70) >consensus CAAUGACAGUAGUCUUGUGAUCAACGUCAAAAGCAAUUGUCAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUU ...(((((((.(.(((.((((....)))).)))).)))))))...(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).. (-42.17 = -42.68 + 0.50)

| Location | 12,975,023 – 12,975,143 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -36.85 |
| Energy contribution | -36.73 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975023 120 - 22407834 AAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUGACAAUUGCUUUUGACGUUGAUCACAAGACUACUGUCAUUG ...........((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))...(((((...((((.(((......))).))).)...)))))... ( -37.20) >DroSec_CAF1 19657 120 - 1 AAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUGACAAUUGCUUUUGACGUUGAUCACAAGACUACUGUCAUUG ...........((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))...(((((...((((.(((......))).))).)...)))))... ( -37.20) >DroSim_CAF1 20820 120 - 1 AAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUGACAAUUGCUUUUGACGUUGAUCACAAGACUACUGUCAUUG ...........((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))...(((((...((((.(((......))).))).)...)))))... ( -37.20) >DroYak_CAF1 22647 119 - 1 AA-AAAAACAAGAGAGAGUGGAGUGGGAGAGUAACUCUCCGCUACGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUGACAAUUGCUUUUGACGUUGAUCACAAGACUACUGUCAUUG ..-........((((((((.(((((((((.....))).))))).).)))))))).......(((((....)))))...(((((...((((.(((......))).))).)...)))))... ( -37.30) >consensus AAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUGACAAUUGCUUUUGACGUUGAUCACAAGACUACUGUCAUUG ...........((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))...(((((...((((.(((......))).))).)...)))))... (-36.85 = -36.73 + -0.12)


| Location | 12,975,063 – 12,975,183 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -31.62 |
| Energy contribution | -32.62 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975063 120 + 22407834 CAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUUUGCUGAUAAUUAUGAAAACCCGCAUAUUUUGAAAAACAUG ......((((((..((((.((((((((((((((...(((.((((((...))))))..)))....))))))))))))))))))........((((........)))).))))))....... ( -33.80) >DroSec_CAF1 19697 120 + 1 CAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUUCGCUGAUAAAUAUGAAAACCCGCAUAUUUUGAAAAACAUG ......(((((...((((.((((((((((((((...(((.((((((...))))))..)))....))))))))))))))))))......((((((........)))))))))))....... ( -36.90) >DroSim_CAF1 20860 120 + 1 CAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUUUGCUGAUAAAUAUGAAAACCCGCAUAUUUUGAAAAACAUG .....(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).........(((((((........)))))))........... ( -35.60) >DroYak_CAF1 22687 118 + 1 CAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCGUAGCGGAGAGUUACUCUCCCACUCCACUCUCUCUUGUUUUU-UUCGCUGAUAAAUAU-AAAACCCGCAUAUUUUAAAAAACAAG ....((((((....))))))(((((.(.(((.((....))((((((...))))))..)))).))))).(((((((((-(........((((((-.........)))))).)))))))))) ( -30.60) >consensus CAAAUAUUCGAACUCGAAUGGAGAGCGAGAGAGCCAGAGCGGAGAGUUGCUCUCCCACUCCACCCUCUCUUGUUUUUCUUCGCUGAUAAAUAUGAAAACCCGCAUAUUUUGAAAAACAUG .....(((((....)))))((((((((((((((...(((.((((((...))))))..)))....)))))))))))))).........(((((((........)))))))........... (-31.62 = -32.62 + 1.00)

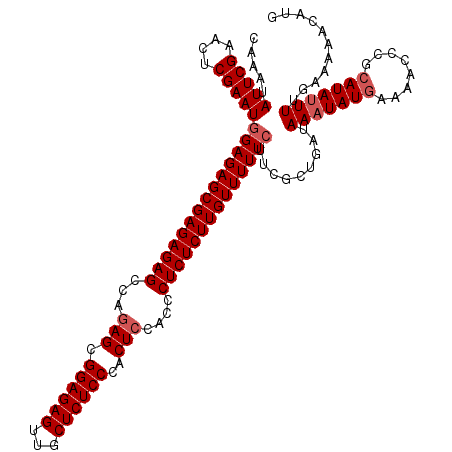
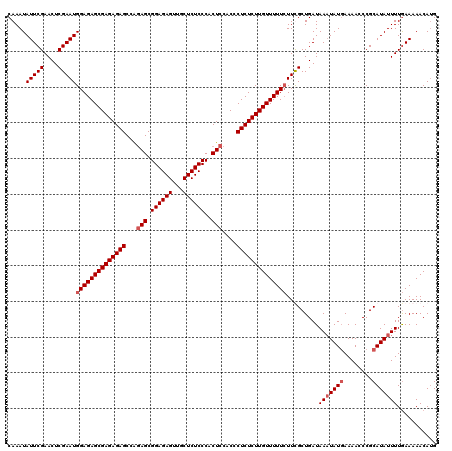
| Location | 12,975,063 – 12,975,183 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -36.65 |
| Energy contribution | -36.52 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975063 120 - 22407834 CAUGUUUUUCAAAAUAUGCGGGUUUUCAUAAUUAUCAGCAAAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUG ..((((((((......(((.(((..........))).)))..)))))))).((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))..... ( -38.20) >DroSec_CAF1 19697 120 - 1 CAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCGAAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUG ..((((((((.(((((((........))))))).((...)).)))))))).((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))..... ( -38.30) >DroSim_CAF1 20860 120 - 1 CAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCAAAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUG ..((((((((......(((.(((..........))).)))..)))))))).((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))..... ( -38.20) >DroYak_CAF1 22687 118 - 1 CUUGUUUUUUAAAAUAUGCGGGUUUU-AUAUUUAUCAGCGAA-AAAAACAAGAGAGAGUGGAGUGGGAGAGUAACUCUCCGCUACGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUG ((((((((((......(((.(((...-......))).))).)-))))))))).((((((((((..((((((...))))))((....)).))).))))))).(((((....)))))..... ( -38.20) >consensus CAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCAAAGAAAAACAAGAGAGGGUGGAGUGGGAGAGCAACUCUCCGCUCUGGCUCUCUCGCUCUCCAUUCGAGUUCGAAUAUUUG ..((((((((......(((.(((..........))).)))..)))))))).((((((((((((((((((.....))).))))))).)))))))).......(((((....)))))..... (-36.65 = -36.52 + -0.13)

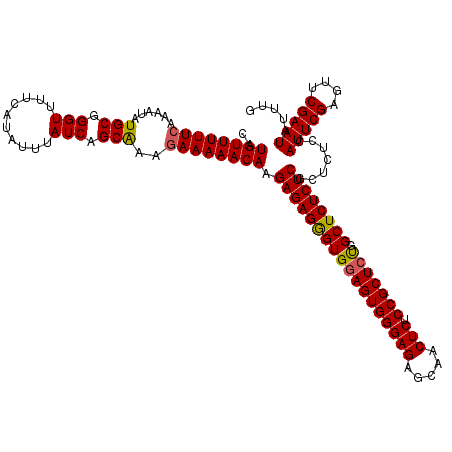
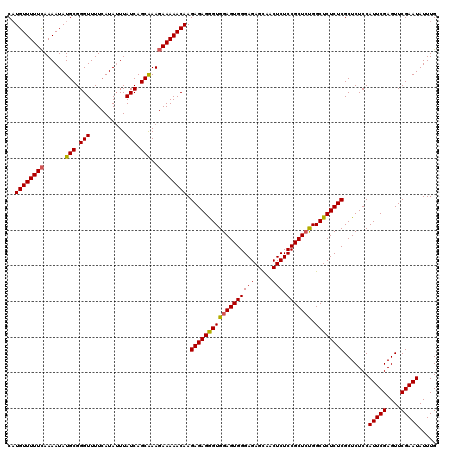
| Location | 12,975,143 – 12,975,263 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12975143 120 - 22407834 AAUCGCUAAAUAAAUAAAAUAAGAGCUGUGAAAAAACAACCACAGGUUGACAAAUUCAACGGAGGAAUGUAACUGAAAUGCAUGUUUUUCAAAAUAUGCGGGUUUUCAUAAUUAUCAGCA ........................((((((((((..(..((.(..(((((.....))))).).))..............((((((((....)))))))))..)))))).......)))). ( -23.21) >DroSec_CAF1 19777 120 - 1 AAUCGCUAAAUAAAUAAAAUAAUAGCUGUGAAAACACACGCACAGGUUAACAAAUUCAACGGAGGAAUGUAACUGAAAUGCAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCG ...((((..(((((((........((((((....)))).))...(((((....((((.......)))).)))))(((((((((((((....)))))))))...)))).))))))).)))) ( -24.90) >DroSim_CAF1 20940 120 - 1 AAUCGCUAAAUAAAUAAAAUAAUAGCUGUGAAAACACACGCACAGGUUAACAAAUUCAACGGAGGAAUGUAACUGAAAUGCAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCA ....(((..(((((((........((((((....)))).))...(((((....((((.......)))).)))))(((((((((((((....)))))))))...)))).))))))).))). ( -24.40) >DroYak_CAF1 22766 119 - 1 AAUCGCAAAAUAAAUAAAAUAAUUACUGUGAAAAUGCACACACAGGUUGACAAAUUCAACGUAGGAAUGUAACUGAAACGCUUGUUUUUUAAAAUAUGCGGGUUUU-AUAUUUAUCAGCG ...(((...................(((((..........)))))(((((.....)))))(...((((((((..((..(((.(((((....))))).)))..))))-))))))..).))) ( -22.50) >consensus AAUCGCUAAAUAAAUAAAAUAAUAGCUGUGAAAACACACGCACAGGUUAACAAAUUCAACGGAGGAAUGUAACUGAAAUGCAUGUUUUUCAAAAUAUGCGGGUUUUCAUAUUUAUCAGCA ....(((..(((((((.........(((((..........)))))((((....((((.......)))).)))).((..(((((((((....)))))))))..))....))))))).))). (-19.36 = -19.68 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:22 2006