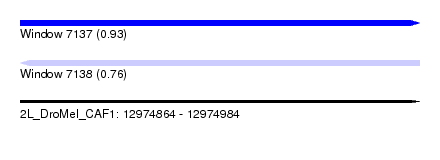
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,974,864 – 12,974,984 |
| Length | 120 |
| Max. P | 0.926463 |

| Location | 12,974,864 – 12,974,984 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974864 120 + 22407834 GCUCGCUUGCUCUCCCAUUUCGAAACUCGAAACAGAAGUGGGAGUUAUUCGUAUUCCGAUAAUGAAAAACCAAUAUGGAGAACGAGCGACGUAAAAAAGGCGGCCCAAAGAUUUUUACCA ((.(((((....(((((((((.............)))))))))((..(((((.((((.(((.((......)).))))))).)))))..)).......)))))))................ ( -34.12) >DroSec_CAF1 19499 119 + 1 GCUCGCUUGCUCUCCCAUUUCGAAACUCGAAACAGAAGUGGGAGUUAUUCGUAUUCCGAUAAUGAAAAACCAAUAUGGAGAACGAGCGACGUAAA-AAGGCGGCCCAAUAAUUUUUACCA ((.(((((....(((((((((.............)))))))))((..(((((.((((.(((.((......)).))))))).)))))..)).....-.)))))))................ ( -34.12) >DroSim_CAF1 20662 119 + 1 GCUCGCUUGCUCUCCCAUUUCGAAACUCGAAACAGAAGUGGGAGUUAUUCGUAUUCCGAUAAUGAAAAACCAAUAUGGAGAACGAGCGACGUAAA-AAGGCGGCCCAAUAAUUUUUACCA ((.(((((....(((((((((.............)))))))))((..(((((.((((.(((.((......)).))))))).)))))..)).....-.)))))))................ ( -34.12) >DroEre_CAF1 8141 103 + 1 GCUCGCUUGCUCUCCCGUUUCGAAACUCCAAACAGAAGUGGAAGUUUUUCGUGCUCCGAUAAUGAAAAUCCAAUAUGGAGAACGAGCGAGUUGAA-AAGGCGGC---------------- (((((((((.(((((.((((.(.....).)))).....((((...(((((((.........)))))))))))....))))).)))))))))....-........---------------- ( -30.90) >DroYak_CAF1 22488 119 + 1 GCUCGCUUGCUCUCCCGUUUAAAAACUCGAAAUACAAGUGGGAGUUUUUCGUAUGCCGAUAAUGAAAAACCAAUAUGGAGAACAAGAGAGCUGAA-AAGGCGCCCCCAAGAUUUUUACCA ((((.((((.(((((..........((((.........)))).(((((((((.........)))))))))......))))).)))).))))((((-((..(........).))))))... ( -26.70) >consensus GCUCGCUUGCUCUCCCAUUUCGAAACUCGAAACAGAAGUGGGAGUUAUUCGUAUUCCGAUAAUGAAAAACCAAUAUGGAGAACGAGCGACGUAAA_AAGGCGGCCCAAAAAUUUUUACCA ((.(((((...((((((((((.............))))))))))...(((((.((((.(((.((......)).))))))).)))))...........)))))))................ (-25.78 = -26.38 + 0.60)



| Location | 12,974,864 – 12,974,984 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.95 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974864 120 - 22407834 UGGUAAAAAUCUUUGGGCCGCCUUUUUUACGUCGCUCGUUCUCCAUAUUGGUUUUUCAUUAUCGGAAUACGAAUAACUCCCACUUCUGUUUCGAGUUUCGAAAUGGGAGAGCAAGCGAGC ..(((((((.....((....)).))))))).(((((.(((((((.((((.((.((((......)))).)).))))............((((((.....)))))).))))))).))))).. ( -35.00) >DroSec_CAF1 19499 119 - 1 UGGUAAAAAUUAUUGGGCCGCCUU-UUUACGUCGCUCGUUCUCCAUAUUGGUUUUUCAUUAUCGGAAUACGAAUAACUCCCACUUCUGUUUCGAGUUUCGAAAUGGGAGAGCAAGCGAGC ..(((((((.....((....))))-))))).(((((.(((((((.((((.((.((((......)))).)).))))............((((((.....)))))).))))))).))))).. ( -33.80) >DroSim_CAF1 20662 119 - 1 UGGUAAAAAUUAUUGGGCCGCCUU-UUUACGUCGCUCGUUCUCCAUAUUGGUUUUUCAUUAUCGGAAUACGAAUAACUCCCACUUCUGUUUCGAGUUUCGAAAUGGGAGAGCAAGCGAGC ..(((((((.....((....))))-))))).(((((.(((((((.((((.((.((((......)))).)).))))............((((((.....)))))).))))))).))))).. ( -33.80) >DroEre_CAF1 8141 103 - 1 ----------------GCCGCCUU-UUCAACUCGCUCGUUCUCCAUAUUGGAUUUUCAUUAUCGGAGCACGAAAAACUUCCACUUCUGUUUGGAGUUUCGAAACGGGAGAGCAAGCGAGC ----------------........-.....((((((.(((((((...(((((.(((((....(((((...(((....)))...)))))..))))).)))))....))))))).)))))). ( -32.20) >DroYak_CAF1 22488 119 - 1 UGGUAAAAAUCUUGGGGGCGCCUU-UUCAGCUCUCUUGUUCUCCAUAUUGGUUUUUCAUUAUCGGCAUACGAAAAACUCCCACUUGUAUUUCGAGUUUUUAAACGGGAGAGCAAGCGAGC ............((..((....))-..))((((.((((((((((.....((((((((.............))))))))...(((((.....))))).........)))))))))).)))) ( -32.42) >consensus UGGUAAAAAUCAUUGGGCCGCCUU_UUUACGUCGCUCGUUCUCCAUAUUGGUUUUUCAUUAUCGGAAUACGAAUAACUCCCACUUCUGUUUCGAGUUUCGAAAUGGGAGAGCAAGCGAGC ...............................(((((.(((((((.((.(((....))).))(((((((.((((................)))).)))))))....))))))).))))).. (-24.63 = -24.95 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:17 2006