
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,974,109 – 12,974,389 |
| Length | 280 |
| Max. P | 0.999943 |

| Location | 12,974,109 – 12,974,229 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -29.70 |
| Energy contribution | -30.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974109 120 - 22407834 GAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAAAGAGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUAAAGAAGUUCGUGGGUUGAUUCUGAAGUUUUGGGAGUAAACGG ....((((((((.(((....(((....))).))).).))))))).(((((...((((.((((......)))).)))).....)))))....(((.(((((.(.....).))))).))).. ( -38.60) >DroSec_CAF1 18761 120 - 1 GAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAGAGCGAGCUAGAGAGCGGGUGGGGUCACACUGGCUUGACGAAGUUCGUGGGUUGAUUCUGAAGUUGUGGGAGUAAACGA ...((((((((..(((....(((....))).)))...))))))))(((((.(.((((.((((......)))).))))..)..)))))....(((.(((((.(.....).))))).))).. ( -42.00) >DroSim_CAF1 19914 120 - 1 GAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAGAGCGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUGACGAAGUUCGUGGGUUGAUUCUGAAGUUCUGGGAGUAAACGG ...((((((((..(((....(((....))).)))...))))))))(((((...((((.((((......)))).)))).....)))))....(((.(((((.(.....).))))).))).. ( -42.20) >DroEre_CAF1 7398 111 - 1 GGAGCUUUCGCCAAUCGAUGUGCGGGAGCGAGACGGAGCGAAACAGAGCUACAGAGCGGGUGGGGUUACACUGGC-AAAAGAAGUUCGUGGGUUGAUUCUGAAGUUAUAGGA-------- ...(((..(((((.....)).)))..)))....((((((((..(((((((.(...((.((((......)))).))-....).))))).))..))).)))))...........-------- ( -35.60) >DroYak_CAF1 21754 120 - 1 GGAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGACGGAGCGAAAGAGAGCUGAAGAGCGGGUGGGGUCACACUGGCCUAAAGAAGUUCGUGGGCUGUUACUGAAGUUUUAGGACUUAACGA ...((((((((((.....)).))))))))..(((((.(((((......((..((.((.((((......)))).))))..))...)))))...)))))....(((((....)))))..... ( -37.20) >consensus GAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAAAGAGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUAAAGAAGUUCGUGGGUUGAUUCUGAAGUUUUGGGAGUAAACGA ....(((((((..(((....(((....))).)))...))))))).(((((...((((.((((......)))).)))).....)))))................................. (-29.70 = -30.42 + 0.72)


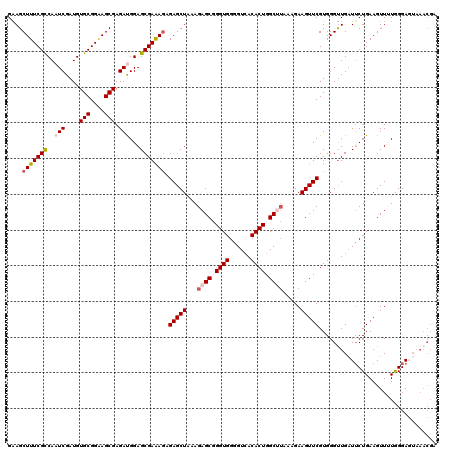
| Location | 12,974,149 – 12,974,269 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -45.98 |
| Consensus MFE | -41.42 |
| Energy contribution | -42.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974149 120 - 22407834 AGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAAAGAGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUAAA .(((((((..(((.(((.......))).)))..)))))))....((((((((.(((....(((....))).))).).))))))).........((((.((((......)))).))))... ( -46.30) >DroSec_CAF1 18801 120 - 1 AGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAGAGCGAGCUAGAGAGCGGGUGGGGUCACACUGGCUUGAC ((((..((((....))))..))))....((((((((.(((...((((((((..(((....(((....))).)))...))))))))..))).....)).((((......)))).)))))). ( -48.50) >DroSim_CAF1 19954 120 - 1 AGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAGAGCGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUGAC ((((..((((....))))..))))....((((((((.(((...((((((((..(((....(((....))).)))...))))))))..))).....)).((((......)))).)))))). ( -48.50) >DroEre_CAF1 7430 119 - 1 AGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGGAGCUUUCGCCAAUCGAUGUGCGGGAGCGAGACGGAGCGAAACAGAGCUACAGAGCGGGUGGGGUUACACUGGC-AAAA .(((((((..(((.(((.......))).)))..)))))))...(((..(((((.....)).)))..))).....(.(((........))).)...((.((((......)))).))-.... ( -42.40) >DroYak_CAF1 21794 120 - 1 AGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGGAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGACGGAGCGAAAGAGAGCUGAAGAGCGGGUGGGGUCACACUGGCCUAAA .(((((((..(((.(((.......))).)))..)))))))....((((((((..((....(((....))).))..).)))))))...(((....)))((((.((......)).))))... ( -44.20) >consensus AGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCGAAAGAGAGCUAAAGAGCGGGUGGGGUCACACUGGCUUAAA .(((((((..(((.(((.......))).)))..)))))))..(((((((((..(((....(((....))).)))...))))....)))))...((((.((((......)))).))))... (-41.42 = -42.18 + 0.76)


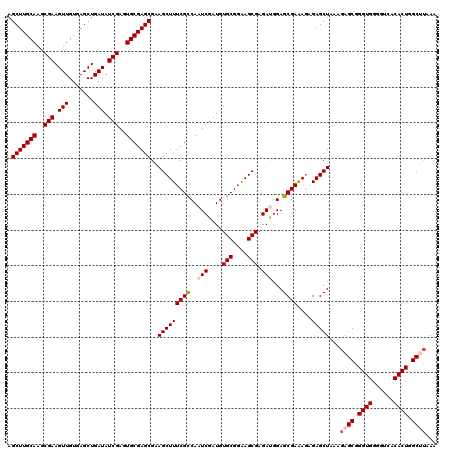
| Location | 12,974,189 – 12,974,309 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -44.40 |
| Energy contribution | -44.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974189 120 - 22407834 UUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCG .(((.(((((....((((....)))).((((.((((....((((..(((......)))..)))).(((((((..(((((.......)))))...)))))))))))))))))))).))).. ( -46.10) >DroSec_CAF1 18841 120 - 1 UUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCAGCGAGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCG .(((.(((((....((((....)))).((((.((((....((((..((((....))))..)))).(((((((.((((((.......)))).)).)))))))))))))))))))).))).. ( -47.40) >DroSim_CAF1 19994 120 - 1 UUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGUCAAUCGAUGUGCGGAAGCGAGAUGGAGCG .(((.(((((....((((....)))).((((.((((....((((..((((....))))..)))).(((((((.((((((.......)))).)).)))))))))))))))))))).))).. ( -47.40) >DroEre_CAF1 7469 120 - 1 UUUCGAUUUCGACCCGUGCGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGGAGCUUUCGCCAAUCGAUGUGCGGGAGCGAGACGGAGCG (((((.(((((.((((..(.((((((.......)))((((((((((((..(((.(((.......))).)))..))))))).......)))))....))))..))))..)))))))))).. ( -49.01) >DroYak_CAF1 21834 120 - 1 UUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGGAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGACGGAGCG (((((.((((...((......((((..((.....)).))))(((((((..(((.(((.......))).)))..))))))))).((((((((((.....)).))))))))))))))))).. ( -45.10) >consensus UUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGCGAAGCUUUCGCCAAUCGAUGUGCGGAAGCGAGAUGGAGCG .(((.(((((....((((....)))).((((.((((....((((..(((......)))..)))).(((((((..(((((.......)))))...)))))))))))))))))))).))).. (-44.40 = -44.40 + -0.00)

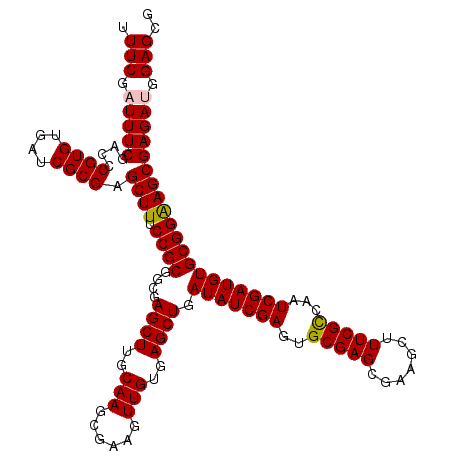

| Location | 12,974,229 – 12,974,349 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -52.18 |
| Consensus MFE | -45.16 |
| Energy contribution | -45.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974229 120 + 22407834 GCUCGCACUCGAUAUCAGCUCACAACUUCGCUUGCAAGCUCGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGAGAGCUUUUCGAAGUGCGAGCGUCGA ((((((((((((....(((((........(((....)))..(((((((........(((((.....)))))...(((....))).)))))))...)))))..))).)))))))))..... ( -49.20) >DroSec_CAF1 18881 120 + 1 GCUCGCACUCGAUAUCAGCUCACAACUUCGGUUGCAAGCUCGCUGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGGGAGCUUUUCGAAGUGCGAGCGUCGA ((((((((((((....(((((.((((....))))....((..((((((........(((((.....)))))...(((....))).))))))..)))))))..))).)))))))))..... ( -52.50) >DroSim_CAF1 20034 120 + 1 GCUCGCACUCGAUAUCAGCUCACAACUUCGGUUGCAAGCUCGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGACUCGCGAUAGGGAGCUUUUCGAAGUGCGAGCGUCGA (((((((((.......((((..((((....))))..)))).....((((((((((.(.(((...((((((((........)))))))).))).).)))))))))).)))))))))..... ( -57.40) >DroEre_CAF1 7509 120 + 1 GCUCGCACUCGAUAUCAGCUCACAACUUCGCUUGCAAGCUCGCCGCGGAAAGCUCGCGAUCGCACGGGUCGAAAUCGAAAUCGAAUCGCGGCAGAGAGCUUUUCGAAGUGCGAGCGUCGA ((((((((((((....(((((........(((....)))..(((((((........(((((.....)))))...(((....))).)))))))...)))))..))).)))))))))..... ( -51.80) >DroYak_CAF1 21874 120 + 1 GCUCGCACUCGAUAUCAGCUCACAACUUCGCUUGCAAGCUCGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGAAGGGAGCUUUUCGAAGUGCGAGCGUCGA ((((((((((((....(((((....(((((.((((.((((..(....)..)))).)))).).(.((..((((........))))..)).))))).)))))..))).)))))))))..... ( -50.00) >consensus GCUCGCACUCGAUAUCAGCUCACAACUUCGCUUGCAAGCUCGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGGGAGCUUUUCGAAGUGCGAGCGUCGA ((((((((((((....(((((........(.((((.((((..(....)..)))).)))).).(.(((.((((........)))).))).).....)))))..))).)))))))))..... (-45.16 = -45.16 + 0.00)

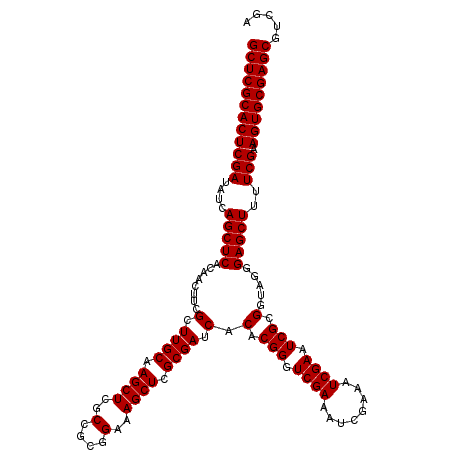
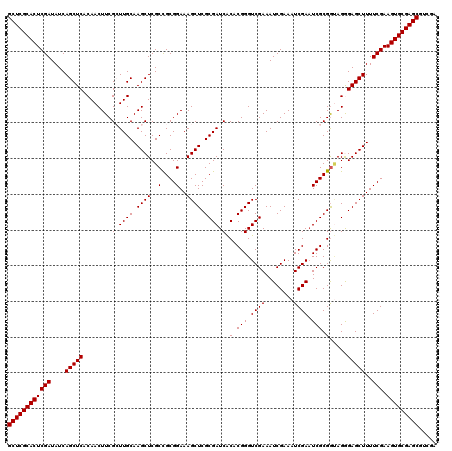
| Location | 12,974,229 – 12,974,349 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -50.82 |
| Consensus MFE | -48.18 |
| Energy contribution | -48.10 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974229 120 - 22407834 UCGACGCUCGCACUUCGAAAAGCUCUCUACCGCGAUUCGAUUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.(((..(((((.(.((((((..((((........))))..)))).....((((((((.(....).)))))))).......)).).)))))....)))))))))))) ( -49.90) >DroSec_CAF1 18881 120 - 1 UCGACGCUCGCACUUCGAAAAGCUCCCUACCGCGAUUCGAUUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCAGCGAGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.(((((((((((...(((((..((((........))))..)))).)...).)))))))........((((..((((....))))..))))....)))))))))))) ( -48.70) >DroSim_CAF1 20034 120 - 1 UCGACGCUCGCACUUCGAAAAGCUCCCUAUCGCGAGUCGAUUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAACCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.((((((((((.(.((((((.(((((........)))))...)))))).).)))))))........((((..((((....))))..))))....)))))))))))) ( -54.00) >DroEre_CAF1 7509 120 - 1 UCGACGCUCGCACUUCGAAAAGCUCUCUGCCGCGAUUCGAUUUCGAUUUCGACCCGUGCGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.(((..(((((.(.((((((..((((........))))..))))((...((((((((.(....).))))))))...))..)).).)))))....)))))))))))) ( -50.80) >DroYak_CAF1 21874 120 - 1 UCGACGCUCGCACUUCGAAAAGCUCCCUUCCGCGAUUCGAUUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.(((..((((((((((((((..((((........))))..)))).....((((((((.(....).))))))))....))))..).)))))....)))))))))))) ( -50.70) >consensus UCGACGCUCGCACUUCGAAAAGCUCCCUACCGCGAUUCGAUUUCGAUUUCGACCCGUGUGAUCGCGAGCUUUCCGCGGCGAGCUUGCAAGCGAAGUUGUGAGCUGAUAUCGAGUGCGAGC .....(((((((((.(((..(((((.(.((((((..((((........))))..)))).....((((((((.(....).)))))))).......)).).)))))....)))))))))))) (-48.18 = -48.10 + -0.08)


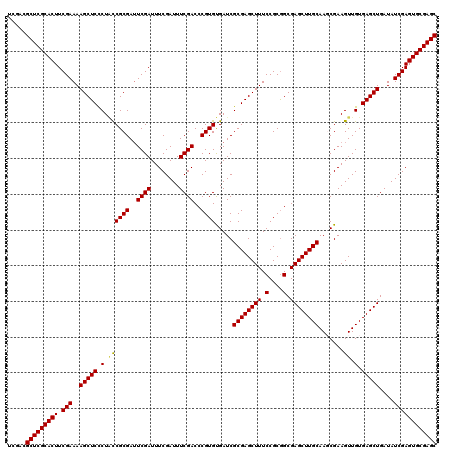
| Location | 12,974,269 – 12,974,389 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -44.22 |
| Consensus MFE | -41.24 |
| Energy contribution | -41.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12974269 120 + 22407834 CGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGAGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .((((((((((((((((((((.....))))..........(((((..((........))..)))))...))))))(....)......)))))))))).(((.(((.......)))..))) ( -41.70) >DroSec_CAF1 18921 120 + 1 CGCUGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGGGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .((((((((((((((.(...(.(.((..((((........))))..)).))..).)))))))))((((((.(((((....))))))))))).))))).(((.(((.......)))..))) ( -40.80) >DroSim_CAF1 20074 120 + 1 CGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGACUCGCGAUAGGGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .((((((((((((((.(.(((...((((((((........)))))))).))).).)))))))))((((((.(((((....))))))))))).))))).(((.(((.......)))..))) ( -49.00) >DroEre_CAF1 7549 120 + 1 CGCCGCGGAAAGCUCGCGAUCGCACGGGUCGAAAUCGAAAUCGAAUCGCGGCAGAGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .(((((((((((((((((.((((((...(((((((((....)))...((........)).)))))).)))))).)).))).))....)))))))))).(((.(((.......)))..))) ( -45.20) >DroYak_CAF1 21914 120 + 1 CGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGAAGGGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .((((((((((((((((((((.....))))....((((((.....((.(....).))...))))))...))))))(....)......)))))))))).(((.(((.......)))..))) ( -44.40) >consensus CGCCGCGGAAAGCUCGCGAUCACACGGGUCGAAAUCGAAAUCGAAUCGCGGUAGGGAGCUUUUCGAAGUGCGAGCGUCGACGUUUUAUUUCUGCGGCUCAGUCGGUUUUAGUUCGUUCUG .((((((((((((((.(.....(.(((.((((........)))).))).)...).)))))))))((((((.(((((....))))))))))).))))).(((.(((.......)))..))) (-41.24 = -41.08 + -0.16)

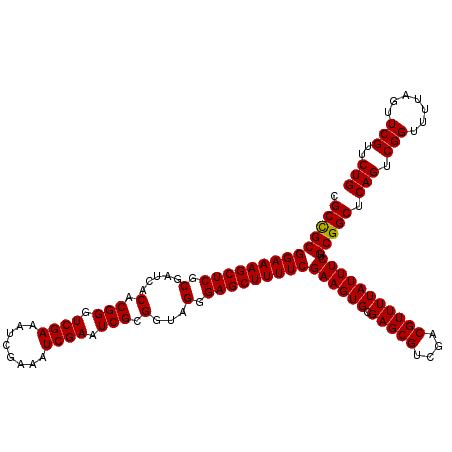
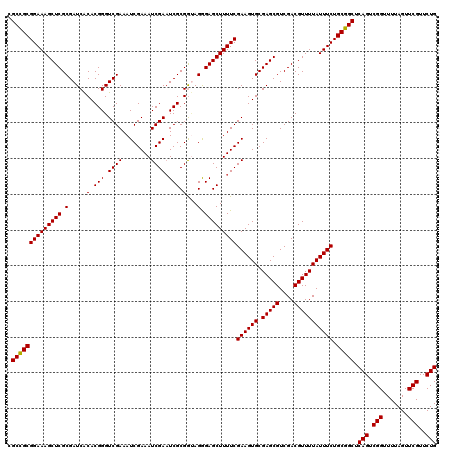
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:15 2006