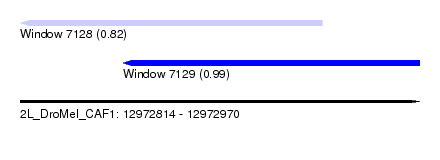
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,972,814 – 12,972,970 |
| Length | 156 |
| Max. P | 0.993052 |

| Location | 12,972,814 – 12,972,932 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.20 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -32.78 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12972814 118 - 22407834 UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCUGCGAUUCAUUGCAAAUUUCUAUCAGCCCUGUGAACGGGGAUUCCCCUCACUGCAGAUGU (((((.((((((((((((((........)))..))))).)))))).)))))((--((((.(((((....)))))............(((((....)))))............)))))).. ( -40.30) >DroSec_CAF1 17646 118 - 1 UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCUGCGAUUCAUUGCAAAUUUCUAUCAGCCCUGGGAACGGGGAAUCCCCUCACUGCAGAUGU (((((.((((((((((((((........)))..))))).)))))).)))))((--((((.(((((....)))))............(((((....)))))............)))))).. ( -40.80) >DroSim_CAF1 18599 118 - 1 UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCAGCGAUUCAUUGCAAAUUUCUAUCAGCCCUGGGAACGGGGAUUCCCCUCACUGCAGAUGU (((((.((((((((((((((........)))..))))).)))))).)))))((--((((..((((....)))).............(((((....)))))............)))))).. ( -40.20) >DroEre_CAF1 6126 118 - 1 UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGCGGCAUGAUGAU--CUGCCAGCGAUUCAUUGGAAAUUUCUAUCAGCUCUGGGAACGGGGAUUCCCCGCACUGCAGAUGU ....(((((((.((....)).......(((((..((((.....))))......--))))).)))))))..((((....))))...((((((((..(((((...)))))..)).)))).)) ( -37.90) >DroYak_CAF1 20232 120 - 1 UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCUGUGAAUGUGGCAUGAGGAUCUCUGCCUGCGAUUCAUUGCAAAUUUCUAUCAGCUCUGCGAACGGGGAUUCCCCGCACUGCAGAUGU ....(((((((((((((..((............))..)))))((((.(((...))))))))))))))).................((((((((..(((((...)))))...)))))).)) ( -41.90) >consensus UCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU__CUGCCUGCGAUUCAUUGCAAAUUUCUAUCAGCCCUGGGAACGGGGAUUCCCCUCACUGCAGAUGU ....(((((((.((....)).......(((((..((((.....))))........))))).))))))).(((((............(((((....)))))...........))))).... (-32.78 = -32.94 + 0.16)


| Location | 12,972,854 – 12,972,970 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -42.66 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12972854 116 - 22407834 CUUAUAAAUCGAUGACGUUGGCAGGCAGCGAG--AGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCUGCGAUUCAUUGCAAAUUUC .........(((((((((.(((((((.((...--.)))).(((((.((((((((((((((........)))..))))).)))))).)))))..--))))).)))..))))))........ ( -42.90) >DroSec_CAF1 17686 113 - 1 CUUAUAAAUCGAUGACGUUGGCAGG---CGAG--AGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCUGCGAUUCAUUGCAAAUUUC .........(((((((((.((((((---(...--.))...(((((.((((((((((((((........)))..))))).)))))).)))))..--))))).)))..))))))........ ( -40.90) >DroSim_CAF1 18639 113 - 1 CUUAUAAAUCGAUGACGUUGGCAGG---CGAG--AGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU--CUGCCAGCGAUUCAUUGCAAAUUUC .........((((((((((((((((---(...--.))...(((((.((((((((((((((........)))..))))).)))))).)))))..--)))))))))..))))))........ ( -44.70) >DroEre_CAF1 6166 115 - 1 CUUAUAAAUCGAUGACGUUGGCAGG---CGAGUGAGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGCGGCAUGAUGAU--CUGCCAGCGAUUCAUUGGAAAUUUC ........(((((((((((((((((---((......)))((((((.((((((((((((((........)))..))))).)))))).))).)))--)))))))))..)))))))....... ( -45.40) >DroYak_CAF1 20272 115 - 1 CUUAUAAAUCGAUGACGUUGGCAGG---CGAG--AGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCUGUGAAUGUGGCAUGAGGAUCUCUGCCUGCGAUUCAUUGCAAAUUUC .........((((((...(.(((((---((((--(...(((.(((..(((((((...(((........))))))))))...))).)))....)))).)))))).).))))))........ ( -39.40) >consensus CUUAUAAAUCGAUGACGUUGGCAGG___CGAG__AGCGCAUCUCGAGUCGCAGCAUUUGCAUAAUAAGGCAGCAGUGCAUGUGGCAUGAGGAU__CUGCCUGCGAUUCAUUGCAAAUUUC .........(((((((((.(((((....((......))..(((((.((((((((((((((........))))..)))).)))))).)))))....))))).)))..))))))........ (-34.36 = -34.60 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:08 2006