
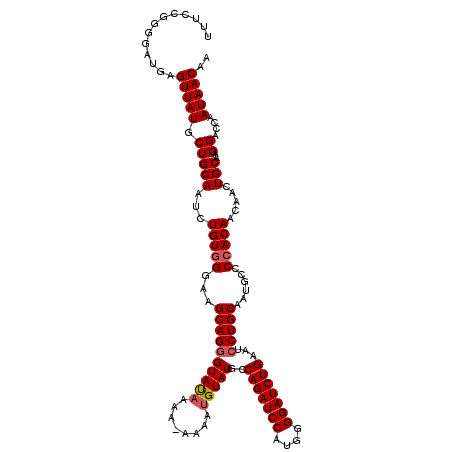
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,969,236 – 12,969,391 |
| Length | 155 |
| Max. P | 0.989756 |

| Location | 12,969,236 – 12,969,356 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12969236 120 + 22407834 AUUCCGGGGAUGAGUUAUGCUGCAAUCUGUGGGAAGCAGGGUACAAAAAAAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCUCAACAACAACUGCAAGGACCAAUAACAA .(((((.((((..((......)).)))).))))).((((((((((........)))))..(((((((....)))))))...)))))..(((............))).............. ( -34.10) >DroSec_CAF1 13943 119 + 1 UUUCCGCGGAUGAGUUAUGCUGCAAUCUGUGGCAAGCAGGGUAUAAAA-AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAAUUGCAAGGACCAAUAACAA ..((((((.........)))((((((.(((((((.((((((((((...-......)))))(((((((....)))))))...)))))..)))).....))))))))).))).......... ( -35.40) >DroSim_CAF1 14761 119 + 1 UUUCCGCGGAUGAGUUAUGCUGCAAUCUGUGGCAAGCAGGGUAUAAAA-AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAACUGCAAGGACCAAUAACAA ..(((((((.((.....(((..(.....)..))).((((((((((...-......)))))(((((((....)))))))...)))))............)).))))..))).......... ( -35.50) >DroEre_CAF1 2654 118 + 1 UUUCCAGGAAUGAGUUAUGCUGCAAUCUGUGGGAAGCAGGGUAUAAAA--AAAUGUAUGCCAGAUCCAUAGGGAUCUGGAUCCUGCAAAGCGCCACAACAACUGCAAGGACCAAUAACAA ..(((.(((.((((.....)).)).)))(((((..((((((((((...--...))))).((((((((....))))))))..)))))....).))))...........))).......... ( -34.90) >DroYak_CAF1 15533 118 + 1 UUACCGGGGAUGAGUUAUGCUGCAAUCUGUGGGAAGCAGGGUAUAAAA--AAAAGUAUGCCAGAUCCAUAGGGAUCUGGAUGCUGCAAUGCCCCACAACAAUUGCAAGGACCCAUAACAA ......(..(((.(((....((((((.((((((..(((..((......--...(((((.((((((((....)))))))))))))))..)))))))))...))))))..))).)))..).. ( -41.60) >consensus UUUCCGGGGAUGAGUUAUGCUGCAAUCUGUGGGAAGCAGGGUAUAAAA_AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAACUGCAAGGACCAAUAACAA .............(((((.(((((...(((((...((((((((((........)))))..(((((((....)))))))...)))))......))))).....)))..))....))))).. (-27.12 = -27.56 + 0.44)


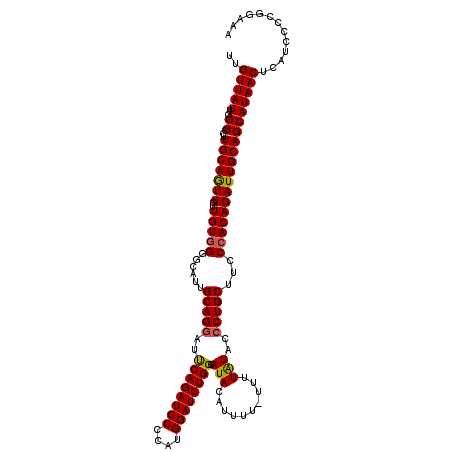
| Location | 12,969,236 – 12,969,356 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -28.86 |
| Energy contribution | -28.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12969236 120 - 22407834 UUGUUAUUGGUCCUUGCAGUUGUUGUUGAGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUUUUUUUGUACCCUGCUUCCCACAGAUUGCAGCAUAACUCAUCCCCGGAAU ......((((....((.((((((.((((......(((((..((((((((....))))))))..((((........)))).))))).((.....))...))))))))))))...))))... ( -30.30) >DroSec_CAF1 13943 119 - 1 UUGUUAUUGGUCCUUGCAAUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU-UUUUAUACCCUGCUUGCCACAGAUUGCAGCAUAACUCAUCCGCGGAAA ..........(((.(((((((..(((..((((..(((((...(((((((....)))))))(....).....-........))))).))))))))))))))((...........))))).. ( -33.60) >DroSim_CAF1 14761 119 - 1 UUGUUAUUGGUCCUUGCAGUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU-UUUUAUACCCUGCUUGCCACAGAUUGCAGCAUAACUCAUCCGCGGAAA ..........(((.(((((((..(((..((((..(((((...(((((((....)))))))(....).....-........))))).))))))))))))))((...........))))).. ( -33.60) >DroEre_CAF1 2654 118 - 1 UUGUUAUUGGUCCUUGCAGUUGUUGUGGCGCUUUGCAGGAUCCAGAUCCCUAUGGAUCUGGCAUACAUUU--UUUUAUACCCUGCUUCCCACAGAUUGCAGCAUAACUCAUUCCUGGAAA ..(((((..((...(((((((..(((((.(....(((((..((((((((....)))))))).(((.....--...)))..)))))..))))))))))))))))))))............. ( -33.30) >DroYak_CAF1 15533 118 - 1 UUGUUAUGGGUCCUUGCAAUUGUUGUGGGGCAUUGCAGCAUCCAGAUCCCUAUGGAUCUGGCAUACUUUU--UUUUAUACCCUGCUUCCCACAGAUUGCAGCAUAACUCAUCCCCGGUAA ..(..((((((..((((((((..(((((((....((((...((((((((....)))))))).(((.....--...)))...)))).)))))))))))))))....))))))..)...... ( -37.30) >consensus UUGUUAUUGGUCCUUGCAGUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU_UUUUAUACCCUGCUUCCCACAGAUUGCAGCAUAACUCAUCCCCGGAAA ..(((((..((...(((((((..(((((......(((((..((((((((....)))))))).(((..........)))..)))))...)))))))))))))))))))............. (-28.86 = -28.62 + -0.24)

| Location | 12,969,276 – 12,969,391 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12969276 115 + 22407834 GUACAAAAAAAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCUCAACAACAACUGCAAGGACCAAUAACAAUGGCAUGAAUAUAACAAAAGCUUCAGUCCCAGA-----CC ...............((((((((((((....))))))...(((((((.((........))..))).))))...........))))))..........................-----.. ( -21.90) >DroSec_CAF1 13983 114 + 1 GUAUAAAA-AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAAUUGCAAGGACCAAUAACAAUGGUAUGAAUAUAACAAAAGCUUCAGUCCCAGA-----CC ........-....(((....(((((((....)))))))..(((((((((...........))))).))))......))).(((..((((............))))...)))..-----.. ( -23.20) >DroSim_CAF1 14801 114 + 1 GUAUAAAA-AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAACUGCAAGGACCAAUAACAAUGGCAUGAAUAUAACAAAAGCUUCAGUCCCAGA-----CC ........-......((((((((((((....))))))...(((((((.((........))..))).))))...........))))))..........................-----.. ( -21.90) >DroEre_CAF1 2694 113 + 1 GUAUAAAA--AAAUGUAUGCCAGAUCCAUAGGGAUCUGGAUCCUGCAAAGCGCCACAACAACUGCAAGGACCAAUAACAAUGGCAUGAAUAUAACAAUAGCUUCAGUCCCAGA-----CC ........--.....((((((((((((....)))))(((.(((((((...............))).))))))).......)))))))..........................-----.. ( -25.26) >DroYak_CAF1 15573 118 + 1 GUAUAAAA--AAAAGUAUGCCAGAUCCAUAGGGAUCUGGAUGCUGCAAUGCCCCACAACAAUUGCAAGGACCCAUAACAAUGGCAUGAAUAUAACAAAAGCUGCAGUCCCAUAAGCAGCC ........--...(((((.((((((((....)))))))))))))(((((...........)))))......((((....))))................(((((..........))))). ( -31.40) >consensus GUAUAAAA_AAAAUGUAUGCCAGAUCCAUGGGGAUCUGAAUCCUGCAAUGCCCCACAACAACUGCAAGGACCAAUAACAAUGGCAUGAAUAUAACAAAAGCUUCAGUCCCAGA_____CC ...............((((((((((((....))))))...(((((((...............))).))))...........))))))................................. (-19.90 = -19.94 + 0.04)


| Location | 12,969,276 – 12,969,391 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12969276 115 - 22407834 GG-----UCUGGGACUGAAGCUUUUGUUAUAUUCAUGCCAUUGUUAUUGGUCCUUGCAGUUGUUGUUGAGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUUUUUUUGUAC ((-----((((((((((((.................((((.......)))).((((((((.(((....))))))))))).))))).))))...))))).....((((........)))). ( -30.30) >DroSec_CAF1 13983 114 - 1 GG-----UCUGGGACUGAAGCUUUUGUUAUAUUCAUACCAUUGUUAUUGGUCCUUGCAAUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU-UUUUAUAC .(-----((((((((..((((...((.(((....))).))..))).)..)))))((((((.((......))))))))))))((((((((....))))))))..........-........ ( -28.20) >DroSim_CAF1 14801 114 - 1 GG-----UCUGGGACUGAAGCUUUUGUUAUAUUCAUGCCAUUGUUAUUGGUCCUUGCAGUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU-UUUUAUAC ((-----((((((((((((.................((((.......)))).((((((((.((......)))))))))).))))).))))...))))).............-........ ( -28.40) >DroEre_CAF1 2694 113 - 1 GG-----UCUGGGACUGAAGCUAUUGUUAUAUUCAUGCCAUUGUUAUUGGUCCUUGCAGUUGUUGUGGCGCUUUGCAGGAUCCAGAUCCCUAUGGAUCUGGCAUACAUUU--UUUUAUAC ((-----((((((((..((((...((.(((....))).))..))).)..)))))(((((.(((....)))..))))))))))(((((((....)))))))..........--........ ( -28.80) >DroYak_CAF1 15573 118 - 1 GGCUGCUUAUGGGACUGCAGCUUUUGUUAUAUUCAUGCCAUUGUUAUGGGUCCUUGCAAUUGUUGUGGGGCAUUGCAGCAUCCAGAUCCCUAUGGAUCUGGCAUACUUUU--UUUUAUAC .(((((..(((...(..((((..((((...(((((((.......)))))))....))))..))))..)..))).)))))..((((((((....)))))))).........--........ ( -38.60) >consensus GG_____UCUGGGACUGAAGCUUUUGUUAUAUUCAUGCCAUUGUUAUUGGUCCUUGCAGUUGUUGUGGGGCAUUGCAGGAUUCAGAUCCCCAUGGAUCUGGCAUACAUUUU_UUUUAUAC ..........(((((((((((...((.(((....))).))..))).))))))))((((((.((......))))))))....((((((((....))))))))................... (-25.46 = -25.42 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:03 2006