
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,968,593 – 12,968,705 |
| Length | 112 |
| Max. P | 0.999452 |

| Location | 12,968,593 – 12,968,705 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
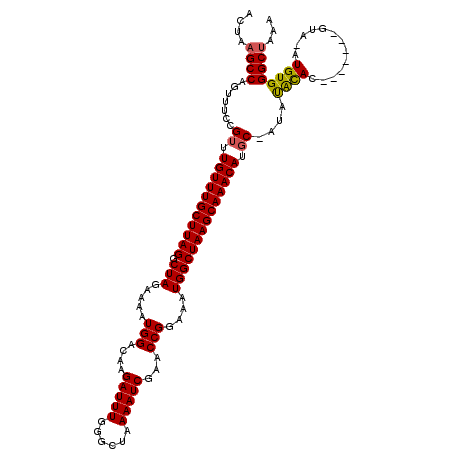
>2L_DroMel_CAF1 12968593 112 + 22407834 UUUAGCCCACAUAUAC-------AUAUGUAU-GCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAAACGGAAACUGGCUUAGU ....((..(((((...-------.)))))..-))......(((.(((((((((((....(((((......)))))..............((....)).......)))))).))))).))) ( -24.60) >DroSec_CAF1 13371 110 + 1 UUUAGCCCACAU--AC-------GUGUAUAU-GCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAACCGGAAACUGGCUUAGU .........((.--((-------((((....-)))))).))((.((((((((((((((.(((((......)))))..............((....))....))))))))).))))).)). ( -29.10) >DroSim_CAF1 14197 110 + 1 UUUAGCCCACAU--AC-------GUGUAUAU-GCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAAACGGAAACUGGCUUAGU .........((.--((-------((((....-)))))).))((.(((((((((((....(((((......)))))..............((....)).......)))))).))))).)). ( -24.60) >DroEre_CAF1 2027 112 + 1 UGUGGCCCACAUGUG-------UGUGUGUGU-GCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAAACGGAAACUGGCUUAGU ....((.(((((...-------...))))).-))......(((.(((((((((((....(((((......)))))..............((....)).......)))))).))))).))) ( -27.70) >DroYak_CAF1 14840 120 + 1 UACAGCCCACAUAUACGAGUAGUAUGCAUGUGGCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAAACGGAAACUGGCUUAGU .(((..((((((((((.....))))..))))))..)))..(((.(((((((((((....(((((......)))))..............((....)).......)))))).))))).))) ( -30.70) >consensus UUUAGCCCACAU_UAC_______GUGUAUAU_GCAUGUUUGCUUAGCCAUUUCCGGUUCGAUUUUAGCCCAAAUCUUGUCCAUUUUCUAGCCUAAGCAAACAAACGGAAACUGGCUUAGU ...((((............................(((((((((((((......))...(((((......)))))................)))))))))))...(....).)))).... (-22.68 = -22.52 + -0.16)



| Location | 12,968,593 – 12,968,705 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -29.34 |
| Energy contribution | -29.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12968593 112 - 22407834 ACUAAGCCAGUUUCCGUUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGC-AUACAUAU-------GUAUAUGUGGGCUAAA ....((((.......((.(((((((((((.(((.....(((....(((((......)))))...)))....)))))))))))))).))-.((((((.-------...))))))))))... ( -35.20) >DroSec_CAF1 13371 110 - 1 ACUAAGCCAGUUUCCGGUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGC-AUAUACAC-------GU--AUGUGGGCUAAA ....(((((.(((((((((......((((.(((((............))))).)))).....)))))))))))))).(((..((((((-........-------))--))))..)))... ( -36.30) >DroSim_CAF1 14197 110 - 1 ACUAAGCCAGUUUCCGUUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGC-AUAUACAC-------GU--AUGUGGGCUAAA ....((((..........(((((((((((.(((.....(((....(((((......)))))...)))....))))))))))))))..(-(((((...-------.)--)))))))))... ( -34.00) >DroEre_CAF1 2027 112 - 1 ACUAAGCCAGUUUCCGUUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGC-ACACACACA-------CACAUGUGGGCCACA .....(((.......((.(((((((((((.(((.....(((....(((((......)))))...)))....)))))))))))))).))-...((((..-------....))))))).... ( -33.50) >DroYak_CAF1 14840 120 - 1 ACUAAGCCAGUUUCCGUUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGCCACAUGCAUACUACUCGUAUAUGUGGGCUGUA ....((((.......((.(((((((((((.(((.....(((....(((((......)))))...)))....)))))))))))))).))(((((..((((.....)))))))))))))... ( -37.00) >consensus ACUAAGCCAGUUUCCGUUUGUUUGCUUAGGCUAGAAAAUGGACAAGAUUUGGGCUAAAAUCGAACCGGAAAUGGCUAAGCAAACAUGC_AUAUACAC_______GUA_AUGUGGGCUAAA ....((((.......((.(((((((((((.(((.....(((....(((((......)))))...)))....)))))))))))))).))....((((.............))))))))... (-29.34 = -29.18 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:59 2006