| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,964,763 – 12,964,881 |
| Length | 118 |
| Max. P | 0.996915 |

| Location | 12,964,763 – 12,964,881 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.53 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
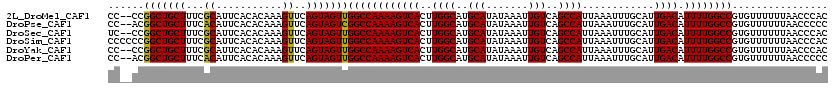
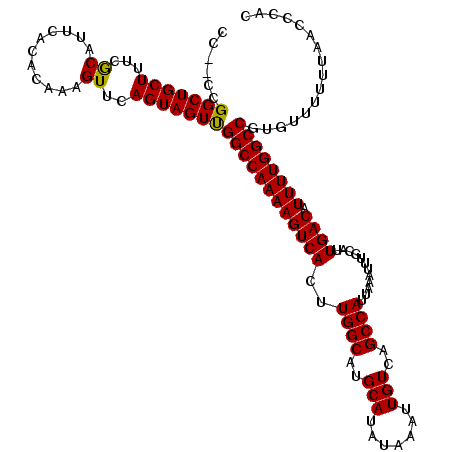
>2L_DroMel_CAF1 12964763 118 + 22407834 GUGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGCGAAAGCAGCCGG--GG (((((((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..))))....(((((.((...(((....))))))))--)) ( -32.24) >DroPse_CAF1 44254 118 + 1 GGGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCGACUACUGAACUUUGUGUGAAUGUGAAAGCAGCCGU--GG .............((((((......(((.(((((.((..(.((.....)).)..)).)))))((((((.....)))))).)))..................(((....))))))))--). ( -28.50) >DroSec_CAF1 9549 118 + 1 GUGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGCGAAAGCAGCCGG--GA ...((((.....((((((((((((.((((............((((((.((.......)))).))))..))))))))))))..(....)......))))....((....))))))..--.. ( -31.74) >DroSim_CAF1 10377 120 + 1 GUGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGCGAAAGCAGCCGGGGGG (((((((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..))))....(((((.((...(((....)))))))))).. ( -33.54) >DroYak_CAF1 11104 118 + 1 GUGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGCGAAAGCAGCCGG--GG (((((((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..))))....(((((.((...(((....))))))))--)) ( -32.24) >DroPer_CAF1 42600 118 + 1 GGGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGUGAAAGCAGCCGU--GG ...((((.....((((((((((((.((((............((((((.((.......)))).))))..))))))))))))..(....)......))))....((....))))))..--.. ( -28.44) >consensus GUGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGUGUGAAUGCGAAAGCAGCCGG__GG ...((((.....((((((((((((.((((............((((((.((.......)))).))))..))))))))))))..(....)......))))....((....))))))...... (-30.19 = -30.64 + 0.45)

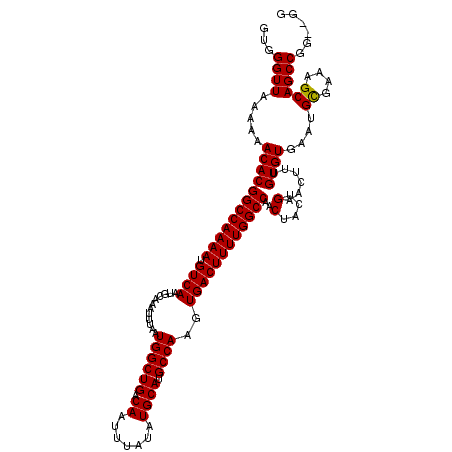
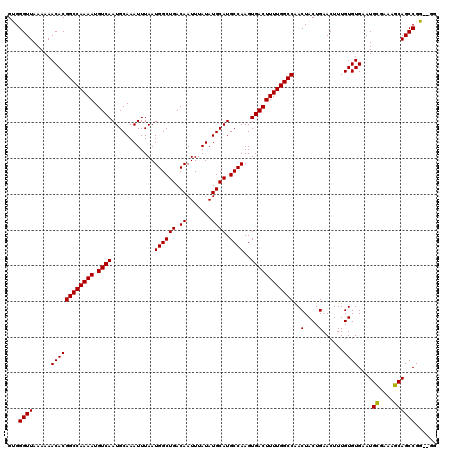
| Location | 12,964,763 – 12,964,881 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.53 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -26.87 |
| Energy contribution | -26.51 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12964763 118 - 22407834 CC--CCGGCUGCUUUCGCAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCAC ..--..(((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).))))))))................ ( -26.14) >DroPse_CAF1 44254 118 - 1 CC--ACGGCUGCUUUCACAUUCACACAAAGUUCAGUAGUCGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCCC .(--(((((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).)))))))))))............. ( -31.24) >DroSec_CAF1 9549 118 - 1 UC--CCGGCUGCUUUCGCAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCAC ..--..(((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).))))))))................ ( -26.14) >DroSim_CAF1 10377 120 - 1 CCCCCCGGCUGCUUUCGCAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCAC ......(((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).))))))))................ ( -26.14) >DroYak_CAF1 11104 118 - 1 CC--CCGGCUGCUUUCGCAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCAC ..--..(((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).))))))))................ ( -26.14) >DroPer_CAF1 42600 118 - 1 CC--ACGGCUGCUUUCACAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCCC .(--(((((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).)))))))))))............. ( -28.44) >consensus CC__CCGGCUGCUUUCGCAUUCACACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCAC ......(((((((...((...........))..)))))))((((((((((((..((((..(((.......)))..))))............)))).))))))))................ (-26.87 = -26.51 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:57 2006